Learning Objectives
●Have a basic understanding of the main interfaces and technologies involved in using Web APIs
●Gain a knowledge of the main chemistry Web APIs
●Be able to pull chemical information programmatically from some chemistry online databases and services
Table of Contents
Module Structure
Week I
8.1 Why using databases programmatically?
8.2 Technological aspects behind using a web API
8.3 Main chemistry APIs
8.2 Technological aspects behind using a web API
One common way to offer open data to be used programmatically is through a web API. Before digging into the most relevant chemistry APIs, we will spend a moment discussing some technological aspects that should be known in order to use it.
What’s a web API?
An API (Application Programming Interface) is the set of elements that a programming library or service makes available to other programmers to be used remotely to access their services and data.
This API is called a web API when these functionalities are delivered via the HTTP protocol through the Internet.
How is a web API used?
In order to be able to get information from an online database, we will need to:
1.Send a query to the database through the Internet (HTTP protocol)
2.Understand the format in which the response will be received
3.Process this response to extract the desired information
Sending the query... - The HTTP protocol and the REST architecture
HTTP stands for HyperText Transfer Protocol and it comprises the set of rules that control the transfer of a resource between a web server and a web client (usually a browser).
The basic elements of an HTTP transaction are:
●A request message
●A response message (which is usually the resource being transferred)
The HTTP request message
The request message consists in some text data sent to an URL (Uniform Resource Locator). A URL is a way of specifying an address in the Internet.
The general structure for an URL consists of
scheme://[user:password@]domain:port/path?query_string#fragment_id
although in HTTP this commonly simplified to
http://domain/path or https://domain/path
The request message, which is usually handled by the HTTP client, has the following structure
●A first line which includes method, path and HTTP protocol version, usually
METHOD /path HTTP/x.x
●Several header lines
●A blank line
●And an optional message body
The HTTP request methods: GET and POST
Although other methods exist, the most common request methods are GET and POST.
In the GET method, any additional information required to specify the request is included in the URL (which means in the path included in the first line of the message). The parameters are included after the path and a question mark as pairs name=value. Parameters are separated by the ampersand symbol (&)
/path?name1=value1&name2=value2
In some cases, as we will see in PubChem API, the parameters are included in the path. This technique is called URL rewriting and shortens the URLs and makes it more usable.
/path/name1/value1/name2/value2
or even
/path/value1/value2
In POST methods, any additional information is not included in the URL but in the body of the message and is usually submitted via a web form. We won’t go further into this method since most of the chemistry APIs can be queried via GET calls.
To end this brief introduction to HTTP, note that some characters are either disallowed or have special meaning in URLs. When these characters appear they will need to be encoded; for example a space is encoded as %20, and a sharp (#) is %23. A reference can be found at http://www.w3schools.com/tags/ref_urlencode.asp.
The HTTP response message
As we have seen for the request, the response contains:
●A first line
●Several header lines
●A blank line
●And a message body which is the transferred resource
Two aspects of the response to be highlighted are the status code (included in the first line) and the content-type information (which is one of the headers).
HTTP status codes are indicated as a three-digit number
●200 means transaction completed successfully
●3xx means some type of redirection
●4xx means a client-side error
○404 means resource not found
●5xx means a server-side error
○500 means unexpected server error
The content type parameter indicates what data is included in the message body. Each type of resource is indicated with a standardized string (Internet media type; aka MIME type). For example, some common response formats are
●HTML file: text/html
●XML file: application/xml or text/xml
●CSV file: text/csv
●JSON file: application/json
●PNG image: image/png
Querying the databases
We have so far seen how the information is transmitted over the WWW. But how are databases accessed over the WWW? There are two main architectures used to access databases over the Net: SOAP and REST. Given its prevalence, we will center our discussion on the REST architecture.
Some major characteristics of the REST (REspresentational State Transfer) architecture are:
●It is usually implemented over HTTP.
●The queries are implemented as GET requests.
●The communication is stateless. Any query must contain all required information. Nothing is saved in the server.|
●The query result is obtained as a response in a predefined format: XML, JSON, CSV, HTML, plain text...
Further reading
●http://code.tutsplus.com/tutorials/http-the-protocol-every-web-developer-must-know-part-1--net-31177
●http://code.tutsplus.com/tutorials/a-beginners-guide-to-http-and-rest--net-16340
●https://www.addedbytes.com/articles/for-beginners/url-rewriting-for-beginners/
●https://www.iana.org/assignments/media-types/media-types.xhtml|
●http://www.restapitutorial.com/
Understanding the responses... - Common structured data file types
Common formats in which a web API may provide its response are HTML, XML, CSV, JSON and plain text. A short explanation of the first four types follows next. Feel free to skip it if you already know their characteristics.
HTML (HyperText Markup Language)
HTML is the standard language used to create web pages. When a web API outputs its result in HTML, this resource is usually intended to be viewed a browser. When no better option is available, HTML can be processed (parsed) to extract a specific information.
Likewise, any web page is published as HTML and so can be processed to extract any relevant information contained therein.
An example of HTML can be found here: http://kinetics.nist.gov/janaf/html/Cl-054.html
This the output of a search in the NIST-JANAF Thermodynamical Tables online database. The full HTML code can be read looking at the source of the page. A fragment is reproduced in Figure 1.
Figure 1. Part of the HTML code of a page
More information on HTML can be found at http://www.w3schools.com/html/.
XML (eXtensible Markup Language)
XML is a markup language designed to contain structured data in format easy to be read by computers and for humans. An example of an XML response can be found running the following query: https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/cid/1/property/MolecularFormula,MolecularWeight,InChIKey/XML. Figure 2 shows its response.
Figure 2. An XML response
More information on XML can be found at http://www.w3schools.com/xml/.
CSV (Comma Separated Values)
CSV, for Comma Separated Values, is a plain-text file format used to store tabular data. In practice, CSV is not a unique format but different variations depending on the implementation and the regional setup of the operating system.
CSV main features:
●Data is stored as plain text.
●Records are separated by new-line characters.
●Each record contains several fields. Columns (fields) are separated by a delimiting character (usually a comma, a semicolon or a tab).
●Each record must have the same number of fields.
CSV main variations:
●Any encoding may be used for the text file: ASCII, Latin-1, UTF-8...
●Values may be quoted or not.
●End-of-line character, delimiting character, quotes and escaping sequences may vary.
●There may be header lines.
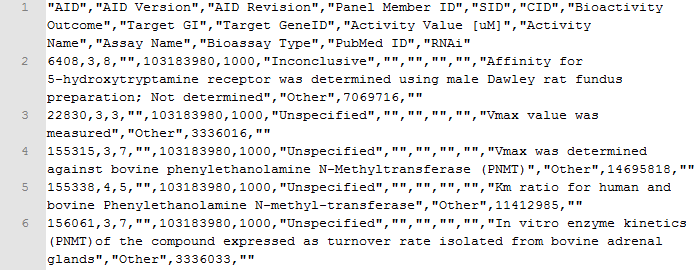
An example of a response in CSV is obtained when running the following query http://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/cid/1000,1001/assaysummary/CSV.
The response is shown in Figure 3.
Figure 3. Fragment of a CSV response
More information can be obtained at https://en.wikipedia.org/wiki/Comma-separated_values.
JSON (JavaScript Object Notation)
JSON, for JavaScript Object Notation, is a format that encodes data object as human-readable text, organized as pairs attribute-value (name:value). Its main features are being a quite compact notation with easy conversion to Javascript objects (eval(), JSON.parse()).
An example query is https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/name/aspirin/property/MolecularWeight/JSON. The response is shown in Figure 4.
Figure 4. A JSON response
More information can be found at http://www.json.org/.
Processing the responses... - Parsers and string functions
Ultimately the response will need to be processed to extract the desired information. This can be done using specific functions or parsers or through the use of some simple string manipulation functions.
Sometimes you want to parse XML or HTML using custom string functions for deleting whitespace, finding the position of certain characters within strings and for extracting substrings. These functions (plus many more) appear in all commonly used languages.
Table 1 summarizes some of these functions in the programming environments we will be discussing next week.
Table 1. Main string functions
|
Spreadsheet (Excel, Calc) |
Basic (VBA, Libre Basic) |
Javascript |
|
SEARCH, FIND |
InStr |
search, indexOf |
|
MID |
Mid |
substr, substring |
|
SUBSTITUTE |
Replace |
replace |
|
TRIM |
Trim |
trim |
|
CONCATENATE |
& (operator), |
concat |
- Log in to post comments
8.3 Main chemistry APIs
CIR (Chemical Identifier Resolver)
The Chemical Identifier Resolver is one of several online tools offered by the CADD Group Chemoinformatics Tools and User Services at the National Cancer Institute.
The CIR web service will convert one identifier (e.g. SMILES) to another (e.g. formula). It can be accessed via a web form with an inbuilt structure editor at http://cactus.nci.nih.gov/chemical/structure or via its web API.
An incomplete web API reference is found at http://cactus.nci.nih.gov/chemical/structure_documentation. Further information can be obtained either by using the form version of the tool or by reading the web services blog: http://cactus.nci.nih.gov/blog.
The general structure for querying the web service is
http://cactus.nci.nih.gov/chemical/structure/”structure identifier”/”representation”
where “structure identifier” is the input and “representation” indicates the desired identifier for output.
An example query is http://cactus.nci.nih.gov/chemical/structure/aspirin/smiles which returns a unique SMILES for aspirin.
Default output format is plain text, but XML output can be obtained by appending /xml to the URL, e.g.http://cactus.nci.nih.gov/chemical/structure/aspirin/names/xml.
OPSIN (Open Parser for Systematic IUPAC nomenclature)
OPSIN (Open Parser for Systematic IUPAC nomenclature) is service provided by the Centre for Molecular Informatics at the University of Cambridge. It can be used to generate a structure (and some common identifiers) from a systematic chemical name.
The reference for the web service can be found at http://opsin.ch.cam.ac.uk/instructions.html. Its entry point ishttp://opsin.ch.cam.ac.uk/opsin.
The query should be set as
http://opsin.ch.cam.ac.uk/opsin/”name”.”type”
where “name” refers to the chemical to be processed and “type” indicates the expected response.
For example, http://opsin.ch.cam.ac.uk/opsin/benzoic+acid.smi returns the SMILES representation of benzoic acid.
CDK (Chemistry Development Kit)
The CDK is an open source program that can generate descriptors from structure. It can be used online as a web API from servers at Uppsala University or Drexel University or by installing in it your own host.
Documentation is available at http://rest.rguha.net/. Servers are provided at the very bottom of the page.
For example, http://ws1.bmc.uu.se:8182/cdk/fingerprint/std/CCO shows a fingerprint for ethanol (plain text response) andhttp://ws1.bmc.uu.se:8182/cdk/depict/200/200/CCO provides a 2D image of this molecule in PNG.
The list of available molecular descriptors can be looked up at /cdk/descriptors.
ChemSpider
ChemSpider APIs allow programmatic access to part of the ChemSpider databases at The Royal Society of Chemistry. They can be accessed through SOAP and/or a REST interface. Some operations require a security token which can be obtained by registering (free) and looking it up in the user profile page.
ChemSpider web services includes four different APIs: Search API, InChI API, MassSpec API and Spectra API. The general documentation is available at http://www.chemspider.com/aboutservices.aspx.
ChemSpider Search API allows searching ChemSpider databases by chemical identifier, structure or properties and retrieve information about the associated records. All operations require a security token and some require a service subscriber role. Not all operations are available through a REST interface. The documentation can be found athttp://www.chemspider.com/Search.asmx.
Response is usually XML and binary information is base64 encoded. Note that some operations are asynchronous. A first call launches the calculation and an id is produced. The id is then used to access the response in a second call to GetAsyncSearchResult.
An example is http://www.chemspider.com/Search.asmx/ GetCompoundInfo?csid=1&token=xxxxxxxx-xxxx-xxxx-xxxx-xxxxxxxxxxxx where the token must be replaced with the user’s one. Results are shown in Figure 5.
Figure 5. A search result from ChemSpider Search API.
ChemSpider InChI API allows interconversion between some chemical identifiers, namely CSID, mol, InChI, InChIKey and SMILES. Few operations require a security token and/or are not available through a REST interface. Documentation is found athttp://www.chemspider.com/InChI.asmx. Response is usually in XML.
An example is http://www.chemspider.com/InChI.asmx/SMILESToInChI?smiles=CCO.
ChemSpider MassSpec API allows searching ChemSpider by formula or by mass. All operations require a security token and some require a service subscriber role. Not all operations are available through a REST interface. Documentation is available athttp://www.chemspider.com/MassSpecAPI.asmx.
Response is usually XML. Some operations are asynchronous; a first call launches the calculation and an id is produced. The id is then used to access the response in a second call to GetAsyncSearchResult (Search API).
An example is the call http://www.chemspider.com/MassSpecAPI.asmx/SearchByMass2?mass=1888&range=0.1.
ChemSpider Spectra API allows searching spectra in ChemSpider database. All operations require a security token with a service subscriber role. The documentation is available at http://www.chemspider.com/Spectra.asmx.
PubChem
PubChem, a product of the National Center for Biotechnology Information, is accessed programmatically through the PUG (Power User Gateway) service. This service can be used either directly or via any of its two interfaces: PUG SOAP or PUG REST. We will, for now, ignore the SOAP interface and focus on the PUG REST service.
When accessed directly, PUG is a queued service that allows searching and downloading of records in the PubChem database: substances and compounds, structures, bioassays... Queries are sent via an HTTP POST call with request and response messages being XML documents. Documentation can be found at https://pubchem.ncbi.nlm.nih.gov/pug/pughelp.html andhttp://www.ncbi.nlm.nih.gov/home/api.shtml.
The PUG REST interface offers a way to access the PubChem database with the convenience of the REST architecture. It allows us to query the PubChem database for data on substances (uncurated data), compounds (curated data) or assays. Documentation is available at https://pubchem.ncbi.nlm.nih.gov/pug_rest/PUG_REST_Tutorial.html andhttps://pubchem.ncbi.nlm.nih.gov/pug_rest/PUG_REST.html. Although most operations are synchronous, some (the asynchronous ones) require a two-step query for obtaining the response (a listkey is returned).
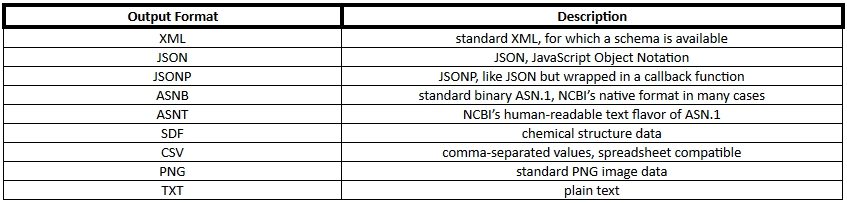
The general structure for using PUG REST is
PUG REST output format can be any of the formats shown in Table 2.
Table 2. PubChem output formats
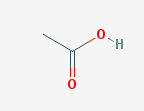
Two examples queries are https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/name/aceticacid/PNG and
https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/name/methanol/property/IUPACName,MolecularWeight/CSV. The responses are shown in Figures 6 and 7.
Figure 6. A PNG response from PubChem PUG REST API.
Figure 7. A CSV response from PubChem PUG REST API.
PDB (Protein DataBank)
RSCB (Research Collaboratory for Structural Bioinformatics) PDB (Protein DataBank) offers three web services to ease accessing to its data. These are documented at http://www.rcsb.org/pdb/software/rest.do.
Currently they include a HTTP POST query method for advanced searching, a simple search REST API for ligands using SMILES as identifier, and a REST API for fetching descriptions of ligands, structures, files, annotations from a PDB id.
The simple search REST API returns the identifiers and descriptions for the ligands matching the search query. For example,http://www.rcsb.org/pdb/rest/smilesQuery?smiles=CC%23C&search_type=substructure performs a substructure search including the group CC#C. Part of the results are shown in Figure 8.
Figure 8. Part of the response to substructure search in PDB.
The fetch API allows the retrieval of the data in the database from its identifiers in PDB. For example,http://www.rcsb.org/pdb/rest/describeHet?chemicalID=CB3 recovers the data for the chemical with id CB3. Responses are commonly in XML.
ChEMBL
The last API accessible database we will consider is ChEMBL, which is a database of bioactive drug-like small molecules hosted at the European Bioinformatics Institute. It provides a RESTful API which responds in XML, JSON or YAML. The main documentation is available at https://www.ebi.ac.uk/chembl/ws.
The API is split into two services:
●data, for accessing the database
●utils, for accessing some cheminformatics tools
The data API allows searching in the ChEMBL database for assays, molecules, target, cell-lines... It includes options for filtering and searching by similarity or substructure. Note that results are paginated. If the number of records returned is larger than 20, you may have to take care of this. This API is documented at https://www.ebi.ac.uk/chembl/api/data/docs.
An example of using this API is
https://www.ebi.ac.uk/chembl/api/data/molecule.json?molecule_structures__canonical_smiles__flexmatch=OCC. The result in JSON format is shown in Figure 10.

Figure 10. JSON response for the ChEMBL data API
The utils API offers some interesting function to obtain molecular descriptors and to generate or process graphical representations of molecules, but requires most inputs as base64 encoded strings. Documentation can be found athttps://www.ebi.ac.uk/chembl/api/utils/docs.
As a side note, you may use https://www.base64encode.org/ to encode and decode base64 strings.
An example use of this utils API is https://www.ebi.ac.uk/chembl/api/data/image/CHEMBL25?format=svg where a SVG image of the molecule with id CHEMBL25 is obtained (see Figure 11).
Figure 11. SVG image obtained from ChEMBL utils API.
- Log in to post comments
Activity 1
Choose a chemical and, using the web services presented in this module, obtain as much information as you can about it. Present the collected information in a report indicating, for each piece the complete URL (parameters included) for the query and the response obtained.
Activity 2
Make a diagram or schema that summarizes which APIs allows you to convert from one information of a chemical to another. At least, the schema should include the following informations: name, IUPAC name, molecular weight, boiling point, SMILES, unique SMILES, mol file, InChI, formula, 2D molecular drawing...
8.4 Other relevant source for data: Other web APIs & web scrapping
Many other data sources are available on the Net. Some of them have APIs to access them programmatically. Some of them can be found in the following directories:
●http://www.programmableweb.com/apis/directory
●https://www.publicapis.com/
●https://app.exiconglobal.com/api-dir/
Wikipedia API
In this module, we will only explore the Wikipedia API, both as an example and as a huge multilingual information source. As you know, Wikipedia is the largest encyclopedia on the Internet. It is free-to-access and is edited by its users.
In terms of processing, we must be aware that it contains semi-structured data which makes it somewhat harder to work with.
Elements of Wikipedia of major interest for a chemist:
●Substance description
●Substance name translations
●Chemboxes (https://en.wikipedia.org/wiki/Template:Chembox)
●Data pages for specific substances: “Properties of” pages, like https://en.wikipedia.org/wiki/Properties_of_water and supplementary data pages: https://en.wikipedia.org/wiki/Aluminium_chloride_%28data_page%29
●Data tables, for example https://en.wikipedia.org/wiki/List_of_elements, https://en.wikipedia.org/wiki/Solubility_table or https://en.wikipedia.org/wiki/Dictionary_of_chemical_formulas
Wikipedia contents can be accessed through the MediaWiki, the wiki engine behind Wikipedia and other projects, API. Its documentation can be accessed at https://www.mediawiki.org/wiki/API:Main_page, https://www.mediawiki.org/wiki/API:Tutorial and https://en.wikipedia.org/w/api.php?action=help&recursivesubmodules=1. A more RESTful API is in development, https://www.mediawiki.org/api/rest_v1/?doc.
The API entry point for each localized Wikipedia is https://LANGUAGE_CODE. wikipedia.org/w/api.php. The language codes for each wikipedia can be looked up at https://www.wikipedia.org/.
The major actions (action=...) to extract data from the MediaWiki API are
- query: to access the data of a page or to look up something in the wikipedia. The major parameters to be set are, prop=revisions, rvprop=content, titles=... and rvcontentformat=...
- parse: to get the processed version of the page. Here the major parameters are prop=text, page=... and contentformat=...
In any case, the response will need to be processed to extract the desired information.
Some example calls are: https://en.wikipedia.org/w/api.php?action=query&prop=revisions&titles=methane&rvprop=content&rvcontentformat=text/x-wiki&format=xml, https://en.wikipedia.org/w/api.php?action=parse&page=ethane%20(data%20page)&prop=text&format=txt.
Other pages at MediaWiki can also accept parameters sent via GET or POST methods. The most important page is index.php. More information can be found at https://en.wikipedia.org/wiki/Help:URL and https://www.mediawiki.org/wiki/Manual:Parameters_to_index.php.
Web scraping
Some information on the web can only be accessed via HTML, that is as a browser does. Even in these cases, the HTML file can be parsed/processed to extract any relevant information. This is called web scraping or also screen scraping.
When making a limited number of accesses, it can be assumed similar to looking up the information with a browser, this does not hold when making a more intensive use of a site. In this later case, one must be aware of the permitted uses of the web site and licenses on the informations you are scraping.
Some example websites with chemical information that could be scraped are listed below:
●Kaye and Laby Online, http://www.kayelaby.npl.co.uk/
●Common Chemistry, http://commonchemistry.org
●MatWeb, http://www.matweb.com
●Solv-DB, http://solvdb.ncms.org/index.html
●eMolecules, https://www.emolecules.com/
●ChemSynthesis, http://www.chemsynthesis.com
●NIST Chemistry Webbook, http://webbook.nist.gov/chemistry/
●Other NIST free access databases, http://srdata.nist.gov/gateway/gateway?dblist=0
●Chemexper, http://www.chemexper.com
●Open Notebook Science, http://onswebservices.wikispaces.com/home
●DrugBank, http://www.drugbank.ca/
●ChemWiki, http://chemwiki.ucdavis.edu
●FDA Substance Registration System, http://fdasis.nlm.nih.gov/srs/srs.jsp
●EMD Millipore Catalog, http://www.emdmillipore.com
●Sigma Aldrich Catalog, http://www.sigmaaldrich.com/
●Alfa Caesar Catalog, https://www.alfa.com/es/catalog/category/chemicals/
●Iowa State University Chemistry Material Safety Data Sheets, http://avogadro.chem.iastate.edu/MSDS/homepage.html
Many more resources are there that can be processed for almost any information that can be needed, including Google (http://www.google.com). In fact, any website that is either static or accepts GET parameters can be used programmatically with the same procedures we will use for web APIs.
- Log in to post comments
8.5 An introduction to programming and to using string functions
We will use this part of the course to introduce a bit of programming and to illustrate the use of the string functions introduced in the first week on the module. Feel free to skip this part if you have experience with programming and processing textual data.
Using strings and string functions in a spreadsheet
In a spreadsheet, strings are typed directly into cells. In fact, any information that cannot be interpreted as a number, a formula, a date or a logical values, is taken to be a string (text).
Any entry can be forced to be interpreted as text by writing a single quote as the first character or by entering it using a formula. In this case the text must be quoted with double quotes.
Figure 1. Main string functions in a spreadsheet.
Figure 1 shows how the different functions are used in a spreadsheet to manipulate textual data. On the left, the formulas are shown; on the right, we can see the results of each formula.
If you have never used these functions, please take some time to replicate Figure 1 and to make yourself familiar to these functions. More information on string functions in spreadsheets can be found at http://www.excelfunctions.net/Excel-Text-Functions.html and https://help.libreoffice.org/Calc/Text_Functions.
A minimal introduction to programming UDF (User Defined Function) in Microsoft Excel for Windows and LibreOffice/OpenOffice Calc
As you may know, spreadsheets can include programming code to extend its functionality. Although different types of programming can be achieved within a spreadsheet, the most common of them is coding user defined functions (UDF). This allows us to create new functions in spreadsheets.
For simple UDF, the flavors of Basic included in Microsoft Excel for Windows and in LibreOffice/OpenOffice are identical. In both cases, the first steps are to open the Basic IDE (Integrated Development Environment) and to add a New Module.
In Microsoft Excel, this is done pressing Alt+F11 and then inserting a module. In LibreOffice/OpenOffice, we will go to the Tools/Macros/Organize Macros/LibreOffice Basic dialog and there add a new module and press Edit.
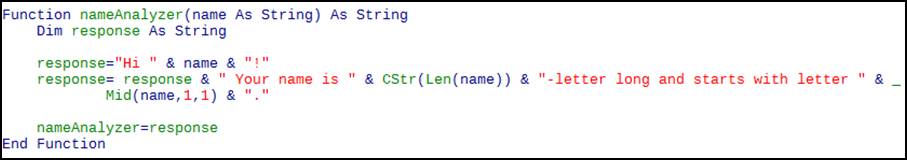
Figure 2. A simple example UDF
Figure 2 shows a simple UDF. Copy it in your module and it will then be usable in formulas in your spreadsheet as the function nameAnalyzer (The name is given in the first line of the code).
More information about Excel VBA and LibreOffice/OpenOffice Basic can be found at https://msdn.microsoft.com/EN-US/library/ee861528.aspx and https://help.libreoffice.org/Basic/Basic_Help.
A minimal introduction to programming in JavaScript
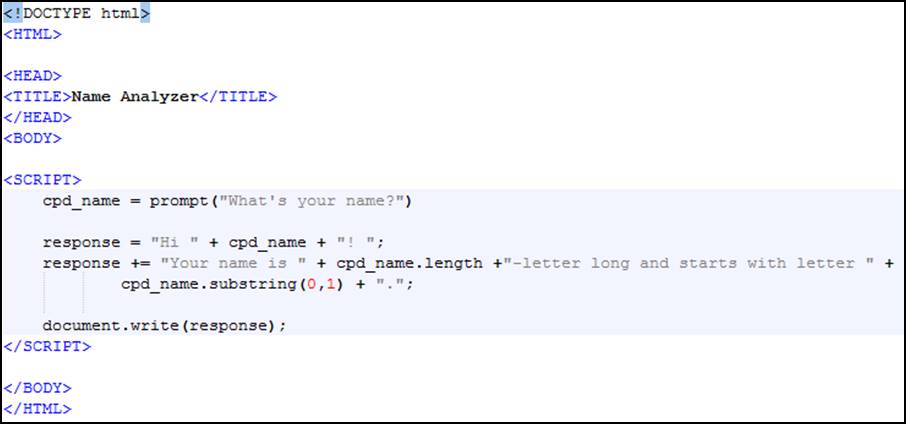
To program a similar functionality in JavaScript (JS), you need to open a text editor (e.g. Notepad on Windows, Vim, LeafPad, Kate or gEdit on Linux, TextEdit on MacOS-X...) and copy the text shown on Figure 3.
Figure 3. A simple example script in JavaScript
As you can see, JavaScript is coded inside an HTML document. Although it can be used in other contexts, JavaScript is one of the key languages of the web with HTML and CSS.
Save your text file using “html” or “htm” for its extensions and open it with a browser. You have coded a HTML/JS page! (A very simple one though).
More information on Javascript is available at http://www.w3schools.com/js/, https://www.codecademy.com/learn/javascript and http://eloquentjavascript.net/.
- Log in to post comments
8.6 Calling the APIs programmatically from desktop and web based applications: Spreadsheets, Javascript & Google scripts
8.6.1: Calling APIs from spreadsheet formulas (no programming)
The simplest way to use a web API from an spreadsheet consist in using the functions HYPERLINK and WEBSERVICE to either create a link to access the information or extract the information itself to further process it.
A minor issue is that the function WEBSERVICE was only recently added to the spreadsheet editors. It is only available from Microsoft Excel 2013 and LibreOffice Calc 4.2. If compatibility with previous versions is required, a Basic based solution will have to be used.
An example file called M8_Example1_Spreadsheet.xlsx is provided in https://drive.google.com/open?id=0B5ln9gFZnHD4V25QMUUzM2Foanc.
8.6.2: Calling APIs from VBA (Visual Basic for Applications), the macro language in Microsoft Office
A web API can also be called from VBA by using several techniques. The two of the simplest options are:
●Calling the worksheet function WEBSERVICE from VBA: . Note this will work only for Microsoft Excel and will be equivalent to the solution just discussed.
●Using an OLE HTTP Client: or . This solution will also work on others flavors of VBA and is compatible with previous editions of Excel.
An example file using the second of this options, M8_Example2_VBA.xlsm is available at https://drive.google.com/open?id=0B5ln9gFZnHD4V25QMUUzM2Foanc.
8.6.3: Calling APIs from LibreOffice/OpenOffice Basic
Similarly, LibreOffice/OpenOffice Basic also offer different ways to query a web API. The simplest option consist in opening a file pipe from an URL by using the Open instruction.
An example is presented in M8_Example3_LOBasic.ods, which can be found at https://drive.google.com/open?id=0B5ln9gFZnHD4V25QMUUzM2Foanc.
Other options like accessing the WEBSERVICE function through the UNO (Universal Network Object) interface or using other programming languages like Python are also possible but won’t be considered here.
8.6.4: Calling APIs from JavaScript
Web APIs can also be called from JavaScript, for example to build online applications. The most common procedures for accessing web services are using the XMLHttpRequest method and the JSONP technique.
XMLHttpRequest is the main interface in Ajax programming. An explanation of this interface can be found at https://developer.mozilla.org/en-US/docs/Web/API/XMLHttpRequest.
An example of querying a web service with the XMLHttpRequest method, M8_Example4_JS_xmlHttpRequest.html, is available
at https://drive.google.com/open?id=0B5ln9gFZnHD4V25QMUUzM2Foanc.
The main limitation of this technology is the same-origin policy enforced by the browsers for security reasons.
The second technique, JSONP (JASON with Padding), is used as an alternative for bypassing the same-origin policy. This technique implies receiving the response as a callback function with a JSON string as an argument. Then this is processed in specially crafted JavaScript function. More information about this technique can be found at http://json-p.org/.
M8_Example5_JS_JSONP.html, available at https://drive.google.com/open?id=0B5ln9gFZnHD4V25QMUUzM2Foanc contains an example of this technology, which is required when calling Wikipedia web API.
We have created a tutorial for you to work through to learn how to access chemical information from Google spreadsheets - gChem. It is available at http://oruopennotebookscience.wikispaces.com/gChem.
- Log in to post comments