CIR (Chemical Identifier Resolver)
The Chemical Identifier Resolver is one of several online tools offered by the CADD Group Chemoinformatics Tools and User Services at the National Cancer Institute.
The CIR web service will convert one identifier (e.g. SMILES) to another (e.g. formula). It can be accessed via a web form with an inbuilt structure editor at http://cactus.nci.nih.gov/chemical/structure or via its web API.
An incomplete web API reference is found at http://cactus.nci.nih.gov/chemical/structure_documentation. Further information can be obtained either by using the form version of the tool or by reading the web services blog: http://cactus.nci.nih.gov/blog.
The general structure for querying the web service is
http://cactus.nci.nih.gov/chemical/structure/”structure identifier”/”representation”
where “structure identifier” is the input and “representation” indicates the desired identifier for output.
An example query is http://cactus.nci.nih.gov/chemical/structure/aspirin/smiles which returns a unique SMILES for aspirin.
Default output format is plain text, but XML output can be obtained by appending /xml to the URL, e.g.http://cactus.nci.nih.gov/chemical/structure/aspirin/names/xml.
OPSIN (Open Parser for Systematic IUPAC nomenclature)
OPSIN (Open Parser for Systematic IUPAC nomenclature) is service provided by the Centre for Molecular Informatics at the University of Cambridge. It can be used to generate a structure (and some common identifiers) from a systematic chemical name.
The reference for the web service can be found at http://opsin.ch.cam.ac.uk/instructions.html. Its entry point ishttp://opsin.ch.cam.ac.uk/opsin.
The query should be set as
http://opsin.ch.cam.ac.uk/opsin/”name”.”type”
where “name” refers to the chemical to be processed and “type” indicates the expected response.
For example, http://opsin.ch.cam.ac.uk/opsin/benzoic+acid.smi returns the SMILES representation of benzoic acid.
CDK (Chemistry Development Kit)
The CDK is an open source program that can generate descriptors from structure. It can be used online as a web API from servers at Uppsala University or Drexel University or by installing in it your own host.
Documentation is available at http://rest.rguha.net/. Servers are provided at the very bottom of the page.
For example, http://ws1.bmc.uu.se:8182/cdk/fingerprint/std/CCO shows a fingerprint for ethanol (plain text response) andhttp://ws1.bmc.uu.se:8182/cdk/depict/200/200/CCO provides a 2D image of this molecule in PNG.
The list of available molecular descriptors can be looked up at /cdk/descriptors.
ChemSpider
ChemSpider APIs allow programmatic access to part of the ChemSpider databases at The Royal Society of Chemistry. They can be accessed through SOAP and/or a REST interface. Some operations require a security token which can be obtained by registering (free) and looking it up in the user profile page.
ChemSpider web services includes four different APIs: Search API, InChI API, MassSpec API and Spectra API. The general documentation is available at http://www.chemspider.com/aboutservices.aspx.
ChemSpider Search API allows searching ChemSpider databases by chemical identifier, structure or properties and retrieve information about the associated records. All operations require a security token and some require a service subscriber role. Not all operations are available through a REST interface. The documentation can be found athttp://www.chemspider.com/Search.asmx.
Response is usually XML and binary information is base64 encoded. Note that some operations are asynchronous. A first call launches the calculation and an id is produced. The id is then used to access the response in a second call to GetAsyncSearchResult.
An example is http://www.chemspider.com/Search.asmx/ GetCompoundInfo?csid=1&token=xxxxxxxx-xxxx-xxxx-xxxx-xxxxxxxxxxxx where the token must be replaced with the user’s one. Results are shown in Figure 5.
Figure 5. A search result from ChemSpider Search API.
ChemSpider InChI API allows interconversion between some chemical identifiers, namely CSID, mol, InChI, InChIKey and SMILES. Few operations require a security token and/or are not available through a REST interface. Documentation is found athttp://www.chemspider.com/InChI.asmx. Response is usually in XML.
An example is http://www.chemspider.com/InChI.asmx/SMILESToInChI?smiles=CCO.
ChemSpider MassSpec API allows searching ChemSpider by formula or by mass. All operations require a security token and some require a service subscriber role. Not all operations are available through a REST interface. Documentation is available athttp://www.chemspider.com/MassSpecAPI.asmx.
Response is usually XML. Some operations are asynchronous; a first call launches the calculation and an id is produced. The id is then used to access the response in a second call to GetAsyncSearchResult (Search API).
An example is the call http://www.chemspider.com/MassSpecAPI.asmx/SearchByMass2?mass=1888&range=0.1.
ChemSpider Spectra API allows searching spectra in ChemSpider database. All operations require a security token with a service subscriber role. The documentation is available at http://www.chemspider.com/Spectra.asmx.
PubChem
PubChem, a product of the National Center for Biotechnology Information, is accessed programmatically through the PUG (Power User Gateway) service. This service can be used either directly or via any of its two interfaces: PUG SOAP or PUG REST. We will, for now, ignore the SOAP interface and focus on the PUG REST service.
When accessed directly, PUG is a queued service that allows searching and downloading of records in the PubChem database: substances and compounds, structures, bioassays... Queries are sent via an HTTP POST call with request and response messages being XML documents. Documentation can be found at https://pubchem.ncbi.nlm.nih.gov/pug/pughelp.html andhttp://www.ncbi.nlm.nih.gov/home/api.shtml.
The PUG REST interface offers a way to access the PubChem database with the convenience of the REST architecture. It allows us to query the PubChem database for data on substances (uncurated data), compounds (curated data) or assays. Documentation is available at https://pubchem.ncbi.nlm.nih.gov/pug_rest/PUG_REST_Tutorial.html andhttps://pubchem.ncbi.nlm.nih.gov/pug_rest/PUG_REST.html. Although most operations are synchronous, some (the asynchronous ones) require a two-step query for obtaining the response (a listkey is returned).
The general structure for using PUG REST is
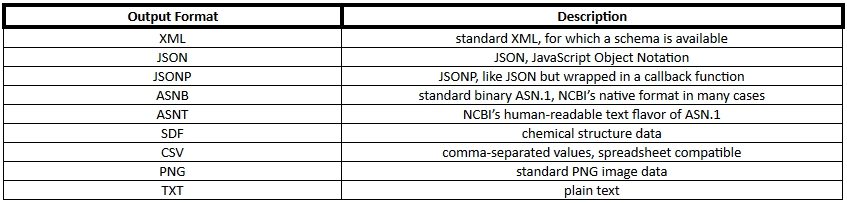
PUG REST output format can be any of the formats shown in Table 2.
Table 2. PubChem output formats
Two examples queries are https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/name/aceticacid/PNG and
https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/name/methanol/property/IUPACName,MolecularWeight/CSV. The responses are shown in Figures 6 and 7.
Figure 6. A PNG response from PubChem PUG REST API.
Figure 7. A CSV response from PubChem PUG REST API.
PDB (Protein DataBank)
RSCB (Research Collaboratory for Structural Bioinformatics) PDB (Protein DataBank) offers three web services to ease accessing to its data. These are documented at http://www.rcsb.org/pdb/software/rest.do.
Currently they include a HTTP POST query method for advanced searching, a simple search REST API for ligands using SMILES as identifier, and a REST API for fetching descriptions of ligands, structures, files, annotations from a PDB id.
The simple search REST API returns the identifiers and descriptions for the ligands matching the search query. For example,http://www.rcsb.org/pdb/rest/smilesQuery?smiles=CC%23C&search_type=substructure performs a substructure search including the group CC#C. Part of the results are shown in Figure 8.
Figure 8. Part of the response to substructure search in PDB.
The fetch API allows the retrieval of the data in the database from its identifiers in PDB. For example,http://www.rcsb.org/pdb/rest/describeHet?chemicalID=CB3 recovers the data for the chemical with id CB3. Responses are commonly in XML.
ChEMBL
The last API accessible database we will consider is ChEMBL, which is a database of bioactive drug-like small molecules hosted at the European Bioinformatics Institute. It provides a RESTful API which responds in XML, JSON or YAML. The main documentation is available at https://www.ebi.ac.uk/chembl/ws.
The API is split into two services:
●data, for accessing the database
●utils, for accessing some cheminformatics tools
The data API allows searching in the ChEMBL database for assays, molecules, target, cell-lines... It includes options for filtering and searching by similarity or substructure. Note that results are paginated. If the number of records returned is larger than 20, you may have to take care of this. This API is documented at https://www.ebi.ac.uk/chembl/api/data/docs.
An example of using this API is
https://www.ebi.ac.uk/chembl/api/data/molecule.json?molecule_structures__canonical_smiles__flexmatch=OCC. The result in JSON format is shown in Figure 10.

Figure 10. JSON response for the ChEMBL data API
The utils API offers some interesting function to obtain molecular descriptors and to generate or process graphical representations of molecules, but requires most inputs as base64 encoded strings. Documentation can be found athttps://www.ebi.ac.uk/chembl/api/utils/docs.
As a side note, you may use https://www.base64encode.org/ to encode and decode base64 strings.
An example use of this utils API is https://www.ebi.ac.uk/chembl/api/data/image/CHEMBL25?format=svg where a SVG image of the molecule with id CHEMBL25 is obtained (see Figure 11).
Figure 11. SVG image obtained from ChEMBL utils API.




Comments 2
Best way to provide multiple identifiers?
Re: Best way to provide multiple identifiers?