Learning Objectives
- Explain what Entrez indices, filters, and links are.
- Explain what depositor-supplied and MeSH synonyms in PubChem are.
- Retrieve compounds that have a particular type of information (e.g., boiling point, melting point, and so on).
- Submit multiple text queries using the Identifier Exchange Service.
- Retrieve annotated information contributed by a given data source.
- Combine multiple queries using Entrez history.
1. PubChem web interfaces for text search
1.1. PubChem Homepage
The PubChem homepage (https://pubchem.ncbi.nlm.nih.gov) provides a search interface that allow users to perform any term/keyword/identifier search against all three major databases of PubChem1-3: Compound, Substance, BioAssay (see Section 3 of Module 4 for data organization in PubChem). If a search returns multiple hits, they are presented on an Entrez DocSum page, which was briefly mentioned in the Module 4 Questions and will also be explained in more detail later in this Module. If the search returns a single record, the user will be directed to the web page that presents information on that record. This page is called the Compound Summary, Substance Record, or BioAssay Record page, depending on the record type (i.e., compound, substance, or assay). In addition, the PubChem homepage provides launch points to various PubChem services, tools, help documents, and more. In general, the PubChem homepage is a central location for all PubChem services.
1.2. Entrez Search and Retrieval System
NCBI’s Entrez4-7 is a database retrieval system that integrates PubChem’s three major databases as well as other NCBI’s major databases, including PubMed, Nucleotide and Protein Sequences, Protein Structures, Genome, Taxonomy, BioSystems, Gene Expression Omnibus (GEO) and many others. Entrez provides users with an integrated view of biomedical data and their relationships. This section focuses on search and retrieval of PubChem data using the Entrez system. A more detailed description on the Entrez system is given in the following documents:
- The Entrez Search and Retrieval System
(http://www.ncbi.nlm.nih.gov/books/NBK184582/) - Entrez Help
(https://www.ncbi.nlm.nih.gov/books/NBK3836/)
1.2.1. Entry points to Entrez
One can search the PubChem databases through Entrez, by initiating a search from the NCBI home page (http://www.ncbi.nlm.nih.gov). By default, if a specific database is not selected in the search menu, Entrez searches all Entrez databases available, and lists the number of records in each database that are returned for this “global query”. The following link directs you to the global query result page for the term “AIDS” against all databases integrated in the Entrez system.
https://www.ncbi.nlm.nih.gov/gquery/?term=AIDS
Simply by selecting one of the three PubChem databases from the global query results page (under the Chemical section), one can see the query results specific to that database.
Alternatively, one can start from the PubChem home page (http://pubchem.ncbi.nlm.nih.gov), where a search of one of the three PubChem databases may be initiated through the search box at the top. It is also possible to initiate an Entrez search against a PubChem database from the following pages:
- https://www.ncbi.nlm.nih.gov/pccompound/ (to search the Compound database)
- https://www.ncbi.nlm.nih.gov/pcsubstance/ (to search the Substance database)
- https://www.ncbi.nlm.nih.gov/pcassay/ (to search the BioAssay database)
1.2.2. Entrez DocSums
If an Entrez search for a query against any of the three PubChem databases returns a single record, the user will be directed to the Compound Summary, Substance Record, or BioAssay Record page for that record (depending on whether the record is a compound, substance, or assay). If it returns multiple records, Entrez will display a document summary report (also called “DocSum” page). The following link directs you to the DocSum page for a search for the term “lipitor” against the PubChem Compound database:
https://www.ncbi.nlm.nih.gov/pccompound?term=lipitor
In this example, the DocSum page displays a list of the compound records returned from the search. For each record, some data-specific information is provided with a link to the summary page for that record. The DocSum page contains controls to change the display type, to sort the results by various means, or to export the page to a file or printer. Additional controls that operate on a query result list are available on the right column of the DocSum page. The DocSum page for the other two PubChem databases look similar to this example for the Compound database.
1.2.3. Entrez Indices
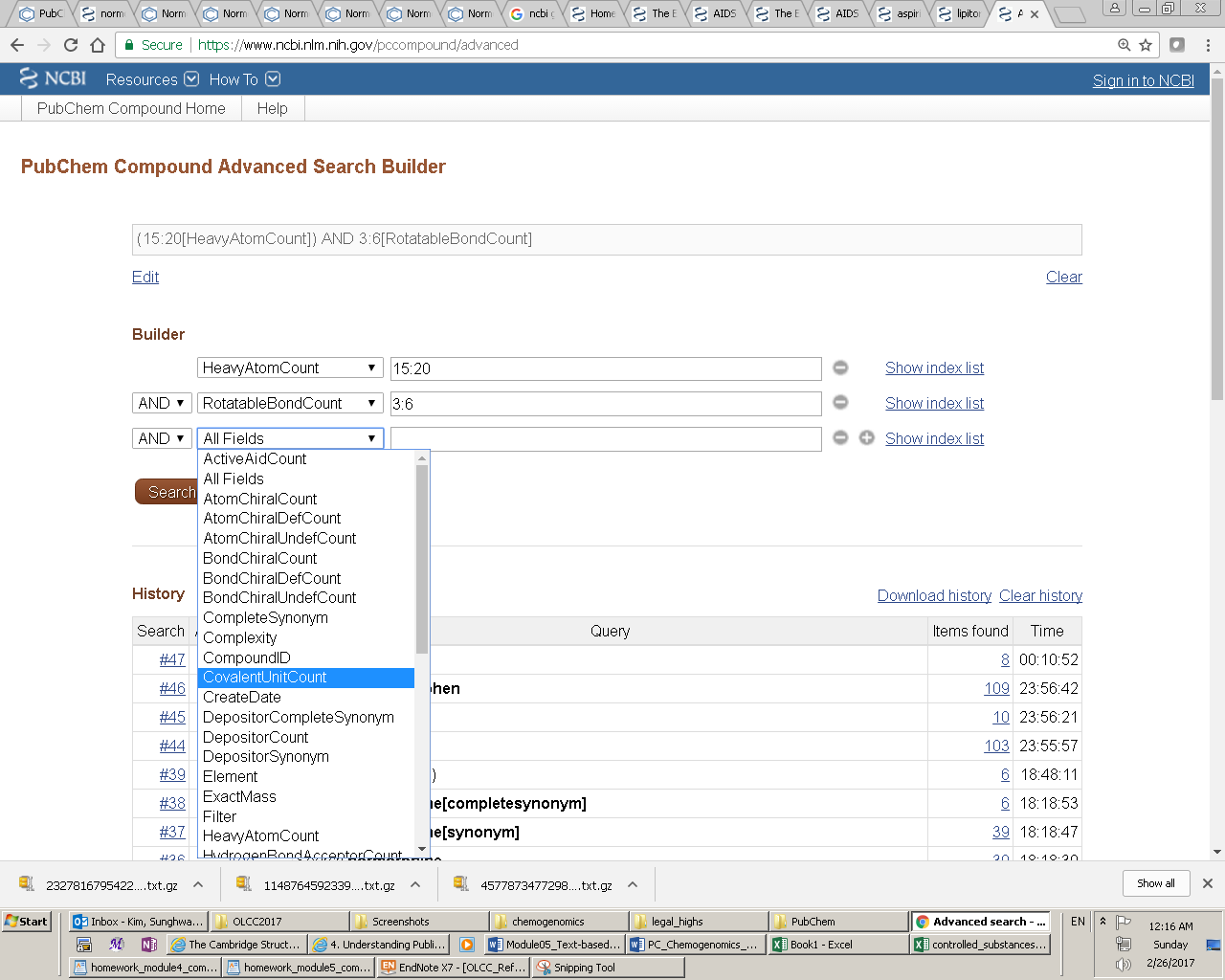
Entrez indices, tied to individual records in an Entrez database, include information on particular aspects (often referred to as fields) of the records. These indices may have text, numeric or date values, and some indices may have multiple values for each record. The available fields and their indexed terms in any Entrez database can be found from the drop-down menus on the Advanced Search Builder page (which can be accessed by clicking the “Advanced” link next to the “Go” button on the PubChem Home page).
When the user enters a query in the Entrez search interface, the Entrez indices are matched directly to that query. By default, in an Entrez search with a simple query, all indexed fields are matched against the query, usually resulting in the largest number of returned records including many unwanted results. One can narrow the search to a particular indexed field, by adding the index name in brackets after the term itself (e.g., “lipitor[synonym]”). For numeric indices, a search for a range of values can be done by using minimum and maximum values separated by a colon and followed by the bracketed index name (e.g., “100:105[MolecularWeight]”). Multiple indices may be searched simultaneously using Entrez’s Boolean operators (e.g., “AND”, “OR” and “NOT”).
A complete list of the Entrez indices available for the three PubChem databases can be retrieved in the XML format, using the eInfo functionality in E-Utilities (which will be covered in Module 7):
- http://eutils.ncbi.nlm.nih.gov/entrez/eutils/einfo.fcgi?db=pccompound (for Compound)
- http://eutils.ncbi.nlm.nih.gov/entrez/eutils/einfo.fcgi?db=pcsubstance (for Substance)
- http://eutils.ncbi.nlm.nih.gov/entrez/eutils/einfo.fcgi?db=pcassay (for BioAssay).
Additional information on the PubChem Entrez indices is available in the “Indices and Filters in Entrez” section of the help documentation:
https://pubchem.ncbi.nlm.nih.gov/help.html#PubChem_Index
1.2.4. Entrez Links
Entrez links are cross links or associations between records in different Entrez databases, or within the same database. These links may be applied to an entire search result list (via the “find related data” section at the right column of a DocSum page) or to an individual record (via links at the bottom of each record presented on the DocSum page). The Entrez links provide a way to discover relevant information in other Entrez databases based on a user’s specific interests. Equivalently, one may think of this as a way to transform an identifier list from one database to another based on a particular criterion. Note that there are limits to how many records may be used as input in a link operation. To process a large amount of input records and/or to expect a large amount of output records associated with the input records, one should use the FLink tool (https://www.ncbi.nlm.nih.gov/Structure/flink/flink.cgi).
A complete list of the Entrez links available for the three PubChem databases can be retrieved in the XML format through these links
- http://eutils.ncbi.nlm.nih.gov/entrez/eutils/einfo.fcgi?db=pccompound (for Compound)
- http://eutils.ncbi.nlm.nih.gov/entrez/eutils/einfo.fcgi?db=pcsubstance (for Substance)
- http://eutils.ncbi.nlm.nih.gov/entrez/eutils/einfo.fcgi?db=pcassay (for BioAssay).
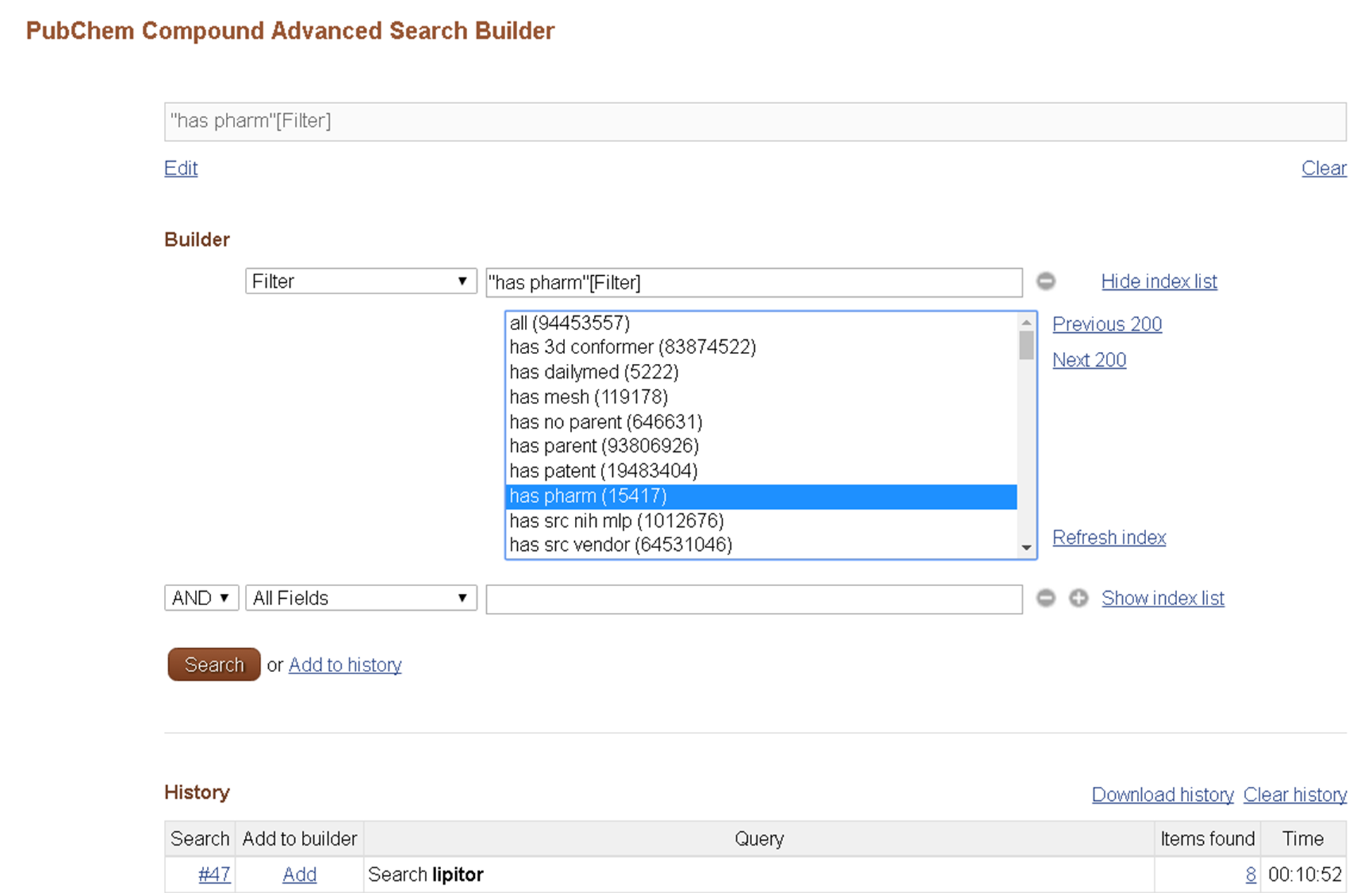
1.2.5. Entrez Filters
Entrez filters are essentially Boolean bits (true or false) for all records in a database that indicate whether or not a given record has a particular property. The Entrez filters may be used to subset other Entrez searches according to this property, by adding the filter to the query string.
Entrez filters are closely related to links in that the majority of Entrez filters in the PubChem databases are generated automatically based on whether PubChem records have Entrez links to a given database. However, some special filters, such as the "lipinski rule of 5" filter, or the “all” filter, are not link-based.
The Entrez filters available for each Entrez database may be found on the Advanced Search Builder page by selecting “Filter” from the “All Fields” dropdown and clicking “Show index list”.
More detailed description of the Entrez filters available for the three PubChem databases are given in the “Indices and Filters in Entrez” section of the help documentation:
https://pubchem.ncbi.nlm.nih.gov/help.html#PubChem_Index
1.2.6. Entrez History
Entrez has a history mechanism (Entrez history) that automatically keeps track of a user’s searches, temporarily caches them (for eight hours), and allows one to combine search result sets with Boolean logic (i.e., “AND”, “OR”, and “NOT”). The Entrez history allows one to limit a search to a subset of records returned from a previous search. Use of Entrez history can help users avoid sending and receiving (potentially) very large lists of identifiers. In addition, through the Entrez history, one can use the search results as an input to various PubChem tools for further manipulation and analysis.
2. Text search in PubChem
2.1. Basics
Text search allows one to find chemical structures using one or more textual keywords, which may be chemical names (e.g., “aspirin”) or any word or phrase that describe molecules of interest (e.g., “cyclooxygenase inhibitors”). One can perform a text search from the PubChem homepage, by providing a text query in the search box. If the query is a phrase or a name with non-alphanumeric characters, double quotes should be used around the query. Various indices can be individually searched by suffixing a text query with an appropriate index enclosed by square brackets (for example, the query “N-(4-hydroxyphenyl)acetamide”[iupacname]). Numeric range searches of appropriate index fields can be performed using a “:” delimiter (for example, the query 100.5:200[molecularweight] for a molecular weight range search between 100.5 and 200.0 g/mol). One can see what search indices are available in PubChem from the drop-down menu on the “PubChem Compound Advanced Search Builder”, which can be accessed by clicking the “advanced” link (next to the “Go” button) on the PubChem homepage. Queries may be combined using the Boolean operators “AND”, “OR”, and “NOT”. These Boolean operators must be capitalized.
2.2. Depositor-supplied synonyms
Conceptually, data in a database are stored in the same way as we would record them in a table or excel spreadsheet. The rows in the table correspond to compounds, and the columns correspond to properties or descriptions for those compounds (e.g., melting and boiling points, chemical names, toxicity, bioactivity, target proteins, and so on). These columns are commonly called “data fields”. You may want to perform a search against all data fields or only a particular field. To search the (depositor-provided) chemical name field of the records in the PubChem Compound database, a chemical name query needs to be suffixed with either of the “[synonym]” or “[completesynonym]” index. The “[synonym]” index invokes search for molecules whose names contain the query chemical name as a part (that is, partial matching), and the “[completesynonym]” index invokes search for those whose names completely match the query (that is, exact matching). If no index is given after the query, PubChem will search all data fields. Compare the following searches for “aspirin” against the PubChem Compound database.
- aspirin[completesynonym] (1 hit, as of Feb. 26, 2017)
https://www.ncbi.nlm.nih.gov/pccompound/?term=aspirin%5Bcompletesynonym%5D - aspirin[synonym] (98 hits)
https://www.ncbi.nlm.nih.gov/pccompound/?term=aspirin%5Bsynonym%5D - aspirin (103 hits)
https://www.ncbi.nlm.nih.gov/pccompound/?term=aspirin
Note that the URLs for these searches contain the query strings (following the string “?term=”), and that the square brackets enclosing the Entrez indices “completesynonym” and “synonym” are replaced with the strings “%5B” and “%5D”. Because the first query resulted in only one hit, the user is directed to the Compound Summary page for the hit compound (CID 2244). On the other hand, because the other two queries result in multiple hits, the results are presented on the DocSum pages.
When either “[completesynonym]” or “[synonym]” is used, it is the “depositor-provided synonyms” fields of the compound records in PubChem that is searched for the query string. The depositor-provided synonyms field for a compound contains a filtered list of chemical names (synonyms) provided by individual data providers for the substances associated with that compound. These synonyms are presented in the “Depositor-provided synonyms” section on a Compound Summary page. To see the variety of synonyms for a compound, check the following link [to the Depositor-provided synonyms” section of the Compound Summary page for CID 2244 (aspirin)]:
https://pubchem.ncbi.nlm.nih.gov/compound/2244#section=Depositor-Supplied-Synonyms
For CID 2244, there are more than 700 depositor-supplied synonyms. These synonyms include not only those commonly used in chemistry class (e.g., common names, IUPAC names, CAS registry numbers) but also those used in many other places (e.g., database identifiers, chemical vendor catalogues, the name of products that contains the chemical, code numbers internally used in a company, and so on).
As mentioned above, the search for aspirin with the “[completesynonym]” index specified returns only one compound (CID 2244). It means that one of many names of this compound exactly matches the query string “aspirin”. On the other hand, the search for aspirin with the “[synonym]” index returns additional 97 compounds. It means that at least one of the names of each these compound partially match the query string (that is, the compound contains the string “aspirin” in one of its names). Interestingly, the results from the last two queries include acetaminophen (CID 1983), which is the active ingredient of Tylenol. Check the following link to the depositor-provided synonyms section of CID 1983 to see what synonyms of Tylenol contains the string “aspirin”:
https://pubchem.ncbi.nlm.nih.gov/compound/1983#section=Depositor-Supplied-Synonyms
Some of the synonyms of Tylenol contains the phrase “aspirin-free” or “non-aspirin”. Note that Tylenol was returned from a search for “aspirin” (through partial matching using the [synonym] index).
2.3. MeSH Synonyms
The National Library of Medicine (NLM)’s Medical Subject Headings (MeSH)8,9
is a controlled vocabulary thesaurus of medical terms arranged in a hierarchical structure. It is used for indexing scientific articles from biomedical journals for PubMed and cataloging medical books, documents, and audiovisual materials, in order to facilitate retrieval of medical information at various levels of specificity.
Many of MeSH terms are chemical names (e.g., for drugs, nutrients, metabolites, toxic chemicals, and so on). PubChem performs an automated annotations of PubChem records with MeSH terms (by means of chemical name matching), creating associations between PubChem records and PubMed articles that share the same MeSH annotation. The MeSH term that match a (depositor-provided) synonym of a compound in PubChem is presented with its entry terms under the “MeSH Synonyms” section of the Compound Summary page of that compound.
Go to the Compound Summary page for CID 171511 via the following link to check the MeSH synonyms and Depositor-supplied Synonyms sections.
https://pubchem.ncbi.nlm.nih.gov/compound/171511#section=Synonyms
CID 171511 (magnyl) is a mixture of aspirin and magnesium oxide. Currently (as of February 2017), the “Depositor-Supplied Synonyms” section of this compound does not have any synonym that contains the string “aspirin”. Therefore, CID 171511 is not returned from a search for aspirin with the [completesynonym] or [synonym] index specified. However, because one of its depositor-provided synonyms, “magnyl” matches the MeSH term “magnyl”, PubChem generate MeSH synonyms for this compound, by annotating it with the MeSH term “magnyl” and its entry terms, which can be found via the following link:
https://meshb.nlm.nih.gov/#/record/ui?ui=C024079
The resulting MeSH synonyms of CID 171511 includes:
- aspirin, magnesium oxide combination
- 2-(acetyloxy)benzoic acid, magnesium oxide mixture
- Acetard
- Magnyl
- aspirin, magnesium oxide mixture
These MeSH synonyms are listed in the “MeSH Synonyms” section of the Compound Summary page for CID 171511. Note that some of the MeSH synonyms now contain the string “aspirin”. When no Entrez index is specified with the query string, “all” indexed fields are searched. Therefore, the search for “aspirin” without the [completesynonym] or [synonym] index does return CID 171511 although no depositor-supplied synonyms contain the string “aspirin”.
3. Additional data retrieval approaches in PubChem
3.1. Classification Browser
The PubChem Classification Browser, which allows the user to navigate or search PubChem records associated to a hierarchical classification system of interest, is available via URL:
http://pubchem.ncbi.nlm.nih.gov/classification
The Classification Browser can also be accessed from the PubChem home page (through the “Services” menu at the top or the “Classification” icon on the right column of the page).
Currently, the Classification Browser can retrieve records annotated with terms in the following classification systems:
- MeSH (Medical Subject Headings)
- ChEBI
- FDA Pharmacological Classification
- KEGG
- LIPID MAPS
- World Health Organization (WHO)’s Anatomical Therapeutic Chemical (ATC)
- World Intellectual Property Organization (WIPO)’s IPC (International Patent Classification)
The Classification Browser provides a powerful way to quickly and visually find a desired subset of PubChem records. The output can be displayed in Tree view or List view.
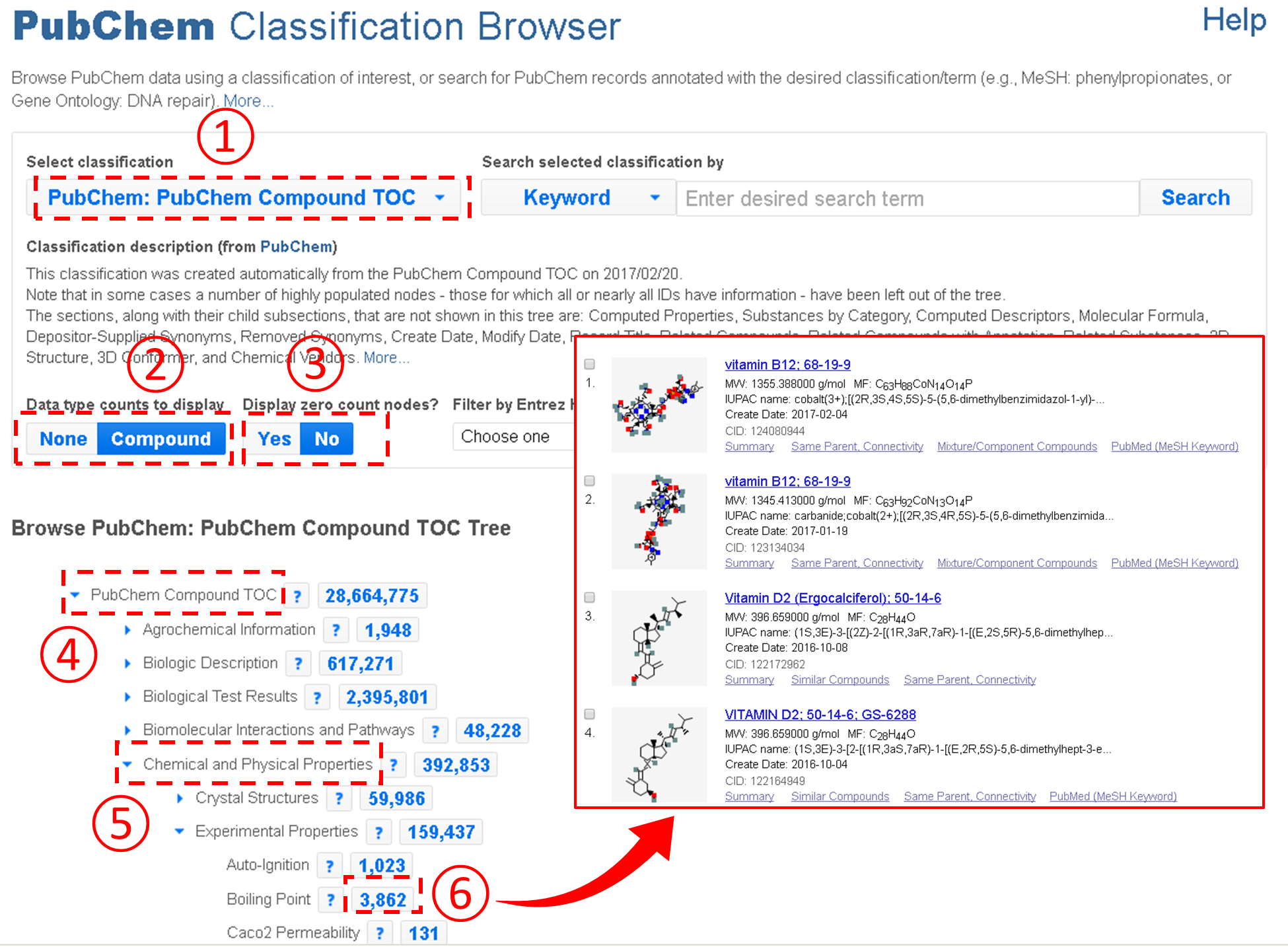
An important feature of the Classification Browser is that the Table of Contents presented on the Compound Summary is integrated into the Classification Browser, allowing users to quickly retrieve compounds with a particular type of information available. For example, the figure below shows how to retrieve all compounds with the boiling point information from PubChem.
In the example above, users need to expand the Table of Contents tree to locate the boiling point node. However, this task may not be easy to some users who do not have prior knowledge about where the node that they want to find is located in the Table of Contents tree system. To assist these users, the Classification Brower supports a keyword search against the node names and descriptions of the classification trees. For example, the example below shows how to retrieve compounds with the CAS Registry number. Note that this task involves a search for the term “CAS”.
The Classification Browser also supports the PubChem BioAssay Classification Tree, providing an additional approach to browse, search, and access the BioAssay data. More detailed information on the Classification Browser is available at the URL:
http://pubchem.ncbi.nlm.nih.gov//classification/docs/classification_help.html
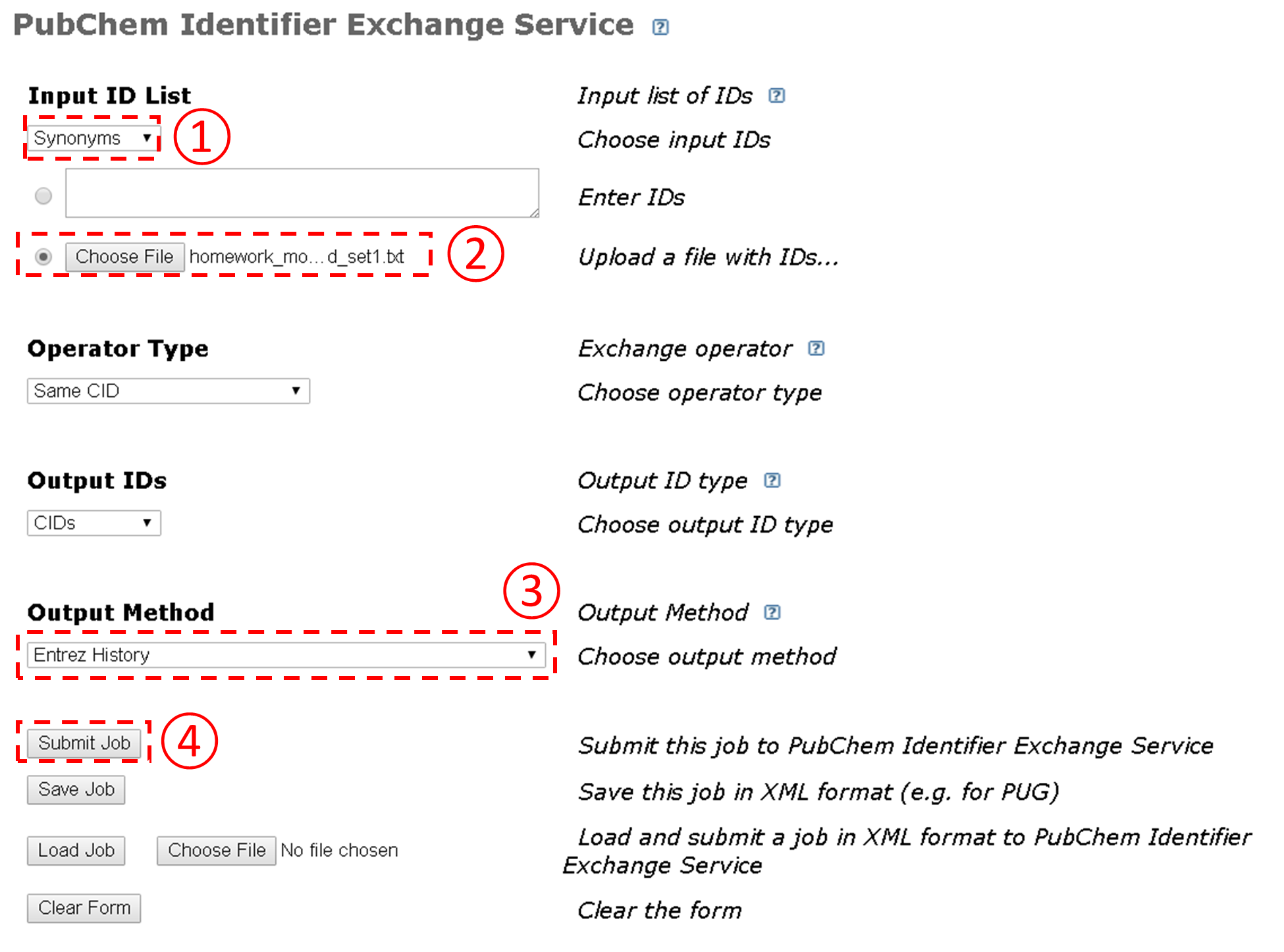
3.2. Identifier Exchange Service
The Identifier Exchange Service can be found at the following URL:
http://pubchem.ncbi.nlm.nih.gov/idexchange
This service allows the user to convert one type of identifiers for a given set of chemical structures into a different type of identifiers for identical or similar chemical structures. Currently, it supports seven types of identifiers: CID, SID, InChI, InChIKey, SMILES, synonyms, Registry ID. When Registry ID is selected as an input or output identifier type, the DSN (Data Source Name) should also be provided.
The input identifier list may be provided using a string, a text file, or Entrez history. When a service request is submitted, it will be queued on PubChem servers. Once the actual task starts to run, the input identifiers will be converted into CIDs (called input CIDs) during the computation, and the CIDs (called output CIDs) that satisfy the condition specified by one of the following operation types will be retrieved:
- Same CID: Same CIDs as input CIDs.
- Same, Stereochemistry: CIDs that have same stereo centers as input CIDs.
- Same, Isotopes: CIDs that have the same isotopes as input CIDs.
- Same, Connectivity: CIDs that have the same connectivity as input CIDs.
- Same parent: CIDs that have the same parents as input CIDs.
- Same parent, Stereochemistry: CIDs that have the same stereo centers and parents as input CIDs.
- Same parent, Isotopes: CIDs that have the same isotopes and parents as input CIDs.
- Same parent, Connectivity: CIDs that have the same connectivity and parents as input CIDs.
- Similar 2D compounds: CIDs similar to the input CIDs in PubChem’s 2-D similarity.
- Similar 3D conformers: CIDs similar to the input CIDs in PubChem’s 3-D similarity.
These output CIDs are then converted into the identifier type specified by the user and written into a file or sent to Entrez history. In practice, the identifier exchange service may be used as a quick approach to search the PubChem Compound database using multiple queries, although this type of task may be performed programmatically (for example, using PUG-REST,10 which will be discussed in Module 7). A more detailed information is available at the URL:
http://pubchem.ncbi.nlm.nih.gov//idexchange/idexchange-help.html
3.3. The PubChem Data Sources page
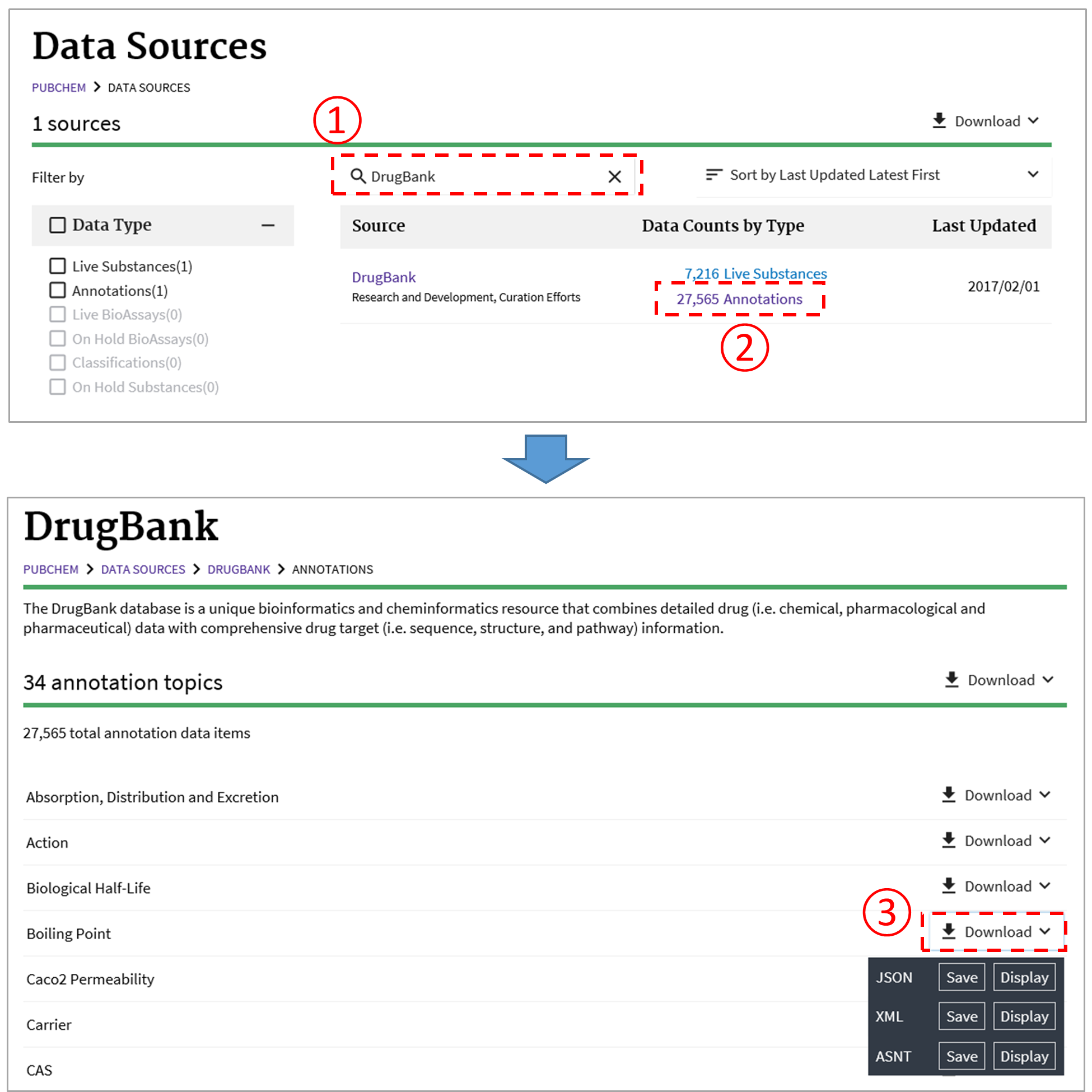
As discussed in Module 4, the PubChem Data Sources page (https://pubchem.ncbi.nlm.nih.gov/sources/) helps users determine who provided what information. This page can be used to retrieve the data provided by a data depositor or to download the annotations collected from a data source. For example, the following figure illustrates how to download the boiling point data collected from DrugBank.11
To obtain a particular kind of annotated information (e.g., boiling points) through the PubChem Data Sources page, one may need to know “in advance” which depositors provide that information. This can be done through a PUG-REST request10 (to be discussed in detail in Module 7). For example, the following PUG-REST request returns all data sources that provide the boiling point information for chemicals.
https://pubchem.ncbi.nlm.nih.gov/rest/pug/annotations/heading/boiling%20point/TXT
On the other hand, one may want to know what kind of information is provided by a given data source. This can also be done using a PUG-REST request:
https://pubchem.ncbi.nlm.nih.gov/rest/pug/annotations/sourcename/DrugBank/TXT
This example retrieves all types of annotations collected from DrugBank.
References
(3) Kim, S. Expert Opinion on Drug Discovery 2016, 11, 843.
(4) Schuler, G. D.; Epstein, J. A.; Ohkawa, H.; Kans, J. A. Methods Enzymol. 1996, 266, 141.
(5) McEntyre, J. Trends in genetics : TIG 1998, 14, 39.
(6) The Entrez Search and Retrieval System (https://www.ncbi.nlm.nih.gov/books/NBK184582/) (Accessed on.
(7) Entrez Help (https://www.ncbi.nlm.nih.gov/books/NBK3836/) (Accessed on.
(8) Medical Subject Headings (MeSH) (https://www.nlm.nih.gov/mesh/) (Accessed on.
(9) Medical Subject Headings (MeSH®) Fact Sheet (https://www.nlm.nih.gov/pubs/factsheets/mesh.html) (Accessed on.
(10) Kim, S.; Thiessen, P. A.; Bolton, E. E.; Bryant, S. H. Nucleic Acids Res. 2015, 43, W605.
Questions
- This question is designed to check if you have a clear understanding of how a text search works in PubChem.
- Go to the PubChem homepage (https://pubchem.ncbi.nlm.nih.gov) and select the “Compound” tab above the search box. Perform three searches using the queries listed on the table below, and record the number of returned compounds and the CIDs of hits.
Query
Number of hits
Returned CIDs
zyrtec[completesynonym]
2
2678, 55182
zyrtec[synonym]
3
2678, 55182, 9850627
zyrtec
5
2678, 55182, 9850627,9551858,5284357
- Find the most frequently occurring covalently-bonded unit in the compounds in the table above. What is the CID of that covalent-bonded unit? [A covalently-bound unit (or simply called covalent unit) consists of a group of covalently-bonded atoms in a compound record. Some compounds in PubChem are mixtures of two or more covalently-bonded units. While the number of components in a mixture is conceptually similar to the number of covalently-bonded units, the term “component” often leads to some ambiguity. For example, is NaCl a single component or a mixture of two components? The use of covalently bonded units (instead of components) removes this ambiguity because it is well accepted that NaCl is bonded through an ionic bond.)]
- What is the CID that is returned from the query “zyrtec[synonym]” but not from “zyrtec[completesynonym]”? Explain why this CID was returned from “zyrtec[synonym]” but not from “zyrtec[completesynonym]”.
- What is the CID that does not have the common covalently-bonded unit in (b)? Explain why it was returned from the query “zyrtec”. [HINT: you will need to compare the depositor-provided synonyms for this CID with the MeSH term “cetirizine” (and its entry terms), which can be accessed through the link below the “MeSH Synonyms” section.]
- Go to the PubChem homepage (https://pubchem.ncbi.nlm.nih.gov) and select the “Compound” tab above the search box. Perform three searches using the queries listed on the table below, and record the number of returned compounds and the CIDs of hits.
- This question tests whether you can search for compounds using molecular property values.
- Read this wikipedia article (https://en.wikipedia.org/wiki/Lipinski's_rule_of_five) and summarize what Lipinski’s rule of 5 is.
- Search PubChem for compounds that satisfy each requirement of Lipinski’s rule of five as well as all the requirements and record the number of compounds in the table below. The queries necessary for these tasks are also given in the table. Note that XLogP is used instead of LogP. (XLogP is a theoretical LogP value predicted by a computer algorithm.) Also note that XLogP has no lower-bound value, while the lowest possible value for the three properties is zero. Currently, the lowest XlogP value in PubChem is -107.5 (for CID 59172357). Therefore, the lower-bound for the XLogP query is set to a sufficiently low value (-1000).
Criteria
Entrez Query
Number of CIDs
#1
HBD 5
0:5[HydrogenBondDonorCount]
#2
HBA 10
0:10[HydrogenBondAcceptorCount]
#3
MW 500
0:500[MolecularWeight]
#4
LogP 5
-1000:5[XLogP]
Compounds satisfying all requirements.
#1 AND #2 AND #3 AND #4
- Read the paper by Congreve et al. (Drug. Discov. Today, 2003, 8(19):876; http://dx.doi.org/10.1016/S1359-6446(03)02831-9) and summarize what Congreve’s rule of 3 is and why it was introduced?
- Based on the table in (b) as a template, make a table that summarizes the number of compounds that satisfy Congreve’s rule of 3. Perform Entrez searches for them and record the number of hits returned.
Criteria
Entrez Query
Number of CIDs
#1
#2
#3
#4
Compounds satisfying all requirements.
- What is the percentage of compounds satisfying Congreve’s rule of 3, relative to all compounds in PubChem?
https://pubchem.ncbi.nlm.nih.gov/compound/6869#section=Biomolecular-Interactions-and-Pathways
On the other hand, some compounds have links to experimental 3-D structures archived in the Cambridge Structural Database (CSD) hosted by the Cambridge Crystallographic Data Centre (CCDC). For example, the 3-D structure of penicillin V archived at CSD-CCDC can be accessed via this URL:
https://pubchem.ncbi.nlm.nih.gov/compound/6869#section=Crystal-Structures
Use PubChem’s classification browser and advanced search builder to find the answer to the following questions.
- Some compounds in PubChem have information on experimentally determined three-dimensional (3-D) molecular structures (presented in the “Protein Bound 3-D Structures” section of the Compound summary page). These structures are provided by the Molecular Modeling Database (MMDB), which curates 3-D structures from Protein Data Bank (PDB). For example, the protein-bound 3-D structure of penicillin V can be accessed via the following URL:
- How many compounds in PubChem have protein-bound 3-D structures?
- How many compounds in PubChem have 3-D structures archived in CSD-CCDC?
- How many compounds in PubChem have both PDB structures and CSD 3-D structures?
- How many compounds in PubChem have any experimental 3-D structures (either from PDB or CSD)?
- What is the ratio of the compounds with both PDB and CSD structures to the compounds with any experimental 3-D structures?
- Explain the difference between PDB (http://www.wwpdb.org/) and CSD (https://www.ccdc.cam.ac.uk/solutions/csd-system/components/csd/).
- Suggest a reason why there are not so many compounds with both PDB and CSD structures.
.
- This question involves the use of PubChem’s Identifier exchange service (https://pubchem.ncbi.nlm.nih.gov/idexchange/idexchange.cgi) to search the Compound database using multiple chemical names as queries.
-
Make a text file that contains the following 10 chemical names:
- 1-(1-Phenylcyclohexyl)pyrrolidine
- 3,4-Methylenedioxymethamphetamine
- Allylprodine
- Barbital
- Cocaine
- Lorazepam
- Methadone
- Normorphine
- oxycodone
- Phenylacetone
- Go to the Identifier Exchange Service, and follow the steps illustrated in the Figure below to search the Compound database using the text file generated from the previous step as an input to the Identifier Exchange Service. How many compounds do you get on the DocSum page?
- Considering that the input file had only ten chemical names, some of them must have resulted in “multiple” hits. To check what chemical name(s) resulted in multiple hits, repeat the search in (b) again, but with the “Output Method” option set to “Two column file showing each input-output correspondence”. What chemical names result in multiple CIDs and what CIDs are associated with them? [In this question, it is not difficult to manually check what they are (because we have only 16 compounds returned from 10 compounds), but it wouldn’t be feasible if you are dealing with hundreds of compounds.]
- Collect the canonical and isomeric SMILES for the CIDs returned in step (c).
- For each chemical name returned in (c), discuss the difference among the multiple compounds (CIDs) returned from the chemical name search.
-
- This question tests whether you know how to obtain desired information from the PubChem Data Sources page (https://pubchem.ncbi.nlm.nih.gov/sources).
- What is the total number of data sources of PubChem information?
- How many data sources does PubChem collect annotations from?
- What kind of annotations does PubChem collect from NCI Investigational Drugs?
- Download the UV data from NCI Investigational Drugs in JSON, and fill in the following table with the information for the compound that appears first in the downloaded file
Compound Name
CID
UV data



Comments 21
Covalently-bonded unit
more information on ChEmBL
Many databases have citation guidelines.
Many databases have citation guidelines at their website. These guidelines are usually found on the "Publications", "Help", "FAQ", or "Citations" page. In the case of ChEMBL, you can find its citation guideline at this URL:
https://www.ebi.ac.uk/chembl/faq#faq43
Covalently-bonded units are ......
Conceptually, covalently-bonded units are components of a compound. Some compound records in PubChem are mixtures or salts, which consist of multiple components, and the concept of covalently-bonded units is used to define the number of components in a compound in PubChem.
Suppose that you have two compounds, CH3COONa (sodium acetate) and CH3COOH (acetic acid). In sodium acetate, the bond between the sodium and oxygen atoms are ionic, so sodium acetate has two covalently-bonded units (sodium and acetate). (Although the sodium atom is not covalently bonded to anything, it is counted as one covalently-bonded unit.) For acetic acid, the H atom of the carboxylic acid group is covalently bonded to the oxygen atom, so acetic acid has one covalently-bonded unit. As another example, the only bond in NaCl is the ionic bond between the Na and Cl atoms, so this compound has two covalently-bonded units.
In your example, all atoms in methanol are connected with covalent bonds, you can view as one covalently-bonded unit (component). However, cetirizine dihydrochloride has three covalently-bonded units (one cetirizine and two hydrochloride). No covalent bonds are formed *between* these three components, so each of them is viewed as a covalent-bonded unit.
Identifier Exchange Service
Split the input into smaller chunks or use FTP
As far as I remember, the ID exchange service can take up to 500,000 CIDs as inputs, but this limit is tightened to 50,000 CIDs if you download images of compounds. If you download more information per compound, you may get a time-out error even if the number of input CIDs does not exceed the limit of ID exchange service.
So, it is recommended to chunk your input list into small pieces and make multiple service requests. If you have much more input CIDs than you can handle in this way, you should download the data from the PubChem ftp site (ftp://ftp.ncbi.nlm.nih.gov/pubchem/).
Can somebody explain me about
The paper has been uploaded.
Thank you for your help.
identifier exchange
Yes, it's been updated.
Hi, Phuc. PubChem's data contents are updated on a daily basis, so it is not unusual to see a small variation in the number of returned records. I've tried the question again and I got the same number of hits as yours. So, consider that the number 16 stated in the question is an approximate number that may vary.
[I added that number as a reference. If a student get an answer that is way off 16, there is a possibility that he or she is doing something wrong. However, if the discrepancy is small, you can consider it's because PubChem's information has been updated.]
Opening JSON file
Opening JSON file
-- DELETED --
-- Deleted, too --
-- Deleted, too --
What is a parent (in PubChem Compound BD)?
Conceptually, it's the important component of a mixture/salt
A "parent" compound is a conceptually important component (part) of a compound. As an example, atorvastatin calcium (lipitor: CID 15378998) has two unique components (atorvastatin and calcium) and we know that it is the atorvastatin part that binds to the target protein, so we view the atorvastatin component is more important than the calcium component.
Of course, the concept of "important components" is very ambiguous, so it needs some clear (mathematical) definition. The current definition of "parent compound" used in PubChem is... if a component contains a super majority (≥70%) of all heavy (non-hydrogen) atoms across all unique components of a mixture and if that component has at least one carbon atom, it is designated as the parent component. In this definition, a single-component (organic) compound is usually considered the parent compound of itself.
A caveat of this definition is that if you have a two-component compound, whose components are similar in size (that is, the heavy atom count ratio of one component to the other is 50:50, no component is considered as a parent.
Is there any charge
Yes, correct.
Some single-component compounds are negatively or positively charged, and their parent compounds are neutralized. So, the acetate ion and acetic acid have the same parent (which is acetic acid).
Entrez filters, links, and indices
Try the following examples:
For Entrez Indices, try Questions 1 and 2 of the Module 5 homework. These questions were designed to help you search PubChem using Entrez indices.
For Entrez filters, try various filters shown in the second Figure on this module (http://olcc.ccce.divched.org/sites/olcc.ccce.divched.org/files/2017OLCCModule5fig2.png). The “has pharm” filter selected in this Figure gives you all compounds that have pharmacological actions. Probably, if you try Question 2, you would feel that getting all compounds satisfying Lipinski’s rule of 5 is somewhat tedious. You can do this using the Entrez filter “Lipinski rule of 5”. (The definition used in this filter is slightly different from those used in the homework question).
For Entrez links, try the dropdown menu called “Find Related Data” on the DocSum page returned from any PubChem search. This dropdown menu is available on the bottom right of the DocSum page. If you are looking for a more practical example, try Homework Question 3 (a) - (d) in Module 6 (not Module 5). You don’t need to read the Module 6 material to solve Question 3(a) - (d). Question 3(d) in Module 6 uses an Entrez Link, but you need to start Question 3(a) to understand the context of the task in 3(d).