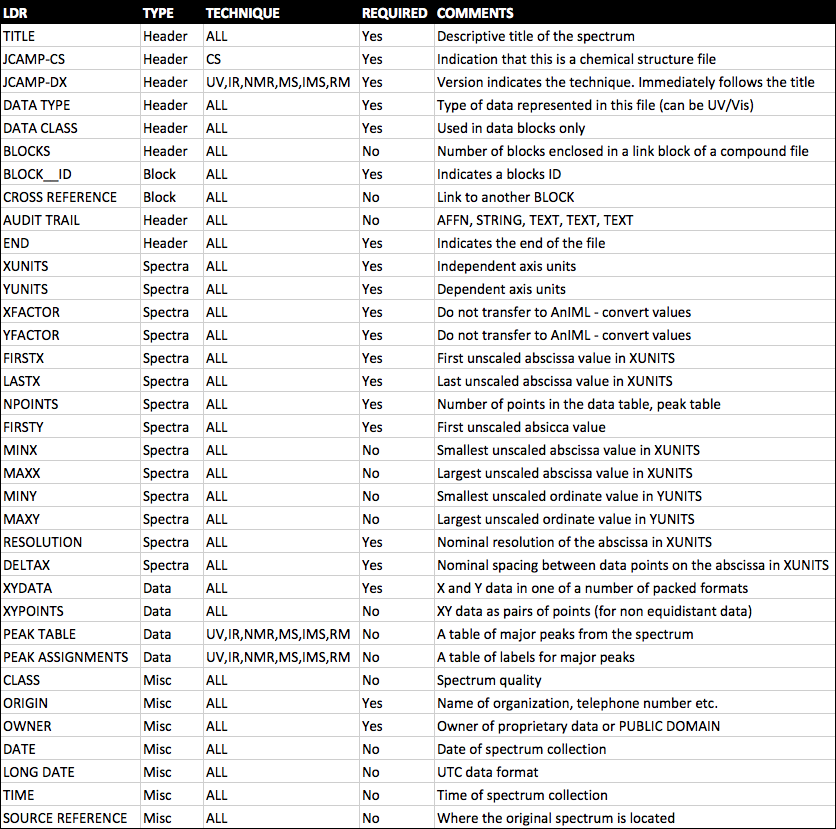
By the end of this module students will:
- Understand the formats for representing spectral data
- JCAMP-DX, AnIML, ANDI, NetCDF, CSV, Tab delimited (XY format)
- Where to obtain reliable spectral information
- AIST Spectral Database for Organic Compounds (SDBS)
- NIST Chemistry WebBook
- ChemSpider
- Simulated spectra
- Spectral software
Module 7: Representing & Managing Digital Spectra (Stuart Chalk)
Since the early 1970’s microcomputers (as they were called at the time) have been a huge part of the development of scientific instrumentation. As computer control of instrumentation became more prevalent, there was a need to also interface the detectors of instruments to the computer so that data (analog or digital) could be captured as it was generated, rather than output it on oscilloscope screens or chart recorders (see https://en.wikipedia.org/wiki/Chart_recorder).
In the early years of the digital capture of spectral data the main limitation was storage capacity. As a result there was a practical limit on the time resolution (points per minute) and signal resolution (how many bits an analog signal was digitized as – see https://en.wikipedia.org/wiki/Analog-to-digital_converter). It wasn’t until the early 1980’s and the advent of the 5 1/4“ floppy disk which initially stored an amazing ~100 kB (0.1 MB) of data, that scientists were easily able to collect and save digital spectra.
Today, instruments generate a vast amount of data and file sizes can be up to several GB each for certain techniques (e.g. GC-MS). This module describes some of the common file formats for spectral data, websites where you can obtain reliable spectral data, and software for viewing/simulating spectra.
7.3: Spectroscopy Software
Jmol with JSpecView - <a href="http://jmol.sourceforge.net/">http://jmol.sourceforge.net/</a> - the best and most widely used online and offline spectral viewer. Browser plugin provides a lot of features for display of data and export in different formats (right mouse click the plugin to see options for viewing spectra and exporting data). Go to the links below to test out the functionality
<a href="http://chemapps.stolaf.edu/jmol/jsmol/jsv.htm">http://chemapps.stolaf.edu/jmol/jsmol/jsv.htm</a> - drag and drop spectra viewing or search for molecules and display simulated spectra
<a href="http://chemapps.stolaf.edu/jmol/jsmol/jsv_jme.htm">http://chemapps.stolaf.edu/jmol/jsmol/jsv_jme.htm</a> - draw a molecule and get simulated
spectrum
<a href="http://chemapps.stolaf.edu/jmol/jsmol/jsv_predict2.htm">http://chemapps.stolaf.edu/jmol/jsmol/jsv_predict2.htm</a> - same as the previous page but with 3D representation of molecular structure
JCAMP Viewer - <a href="http://pslc.uwsp.edu/Viewers.shtml">http://pslc.uwsp.edu/Viewers.shtml</a> - web page looks old but software has been updated recently
Flot JCAMP Viewer - <a href="http://webbook.nist.gov/chemistry">http://webbook.nist.gov/chemistry</a> - get from any page with spectra – requires web server
SpeckTackle Javascript Viewer - <a href="https://bitbucket.org/sbeisken/specktackle">https://bitbucket.org/sbeisken/specktackle</a> - new on the scene – requires web server
OpenSpectralWorks - <a href="http://scanedit.sourceforge.net/">http://scanedit.sourceforge.net/</a>
SpekWin32 - <a href="http://effemm2.de/spekwin/index_en.html">http://effemm2.de/spekwin/index_en.html</a>
OpenChrom - <a href="https://www.openchrom.net/">https://www.openchrom.net/</a> - primarily for chromatographic data but good for mas spectrometry as well
ACD/Labs NMR Processor - <a href="http://www.acdlabs.com/resources/freeware/nmr_proc/index.php">http://www.acdlabs.com/resources/freeware/nmr_proc/index.php</a> - NMR Only
MestreLabs MNova - <a href="http://mestrelab.com/software/mnova/">http://mestrelab.com/software/mnova/</a> - NMR Only ($)
ChemDoodle - <a href="https://www.chemdoodle.com/">https://www.chemdoodle.com/</a> - reads all JCAMP files ($)
Spectral Prediction
nmrdb.org - <a href="http://www.nmrdb.org/">http://www.nmrdb.org/</a>
NMR Shift DB Online - <a href="http://nmrshiftdb.nmr.uni-koeln.de/nmrshiftdb/">http://nmrshiftdb.nmr.uni-koeln.de/nmrshiftdb/</a>
ChemDoodle - <a href="https://web.chemdoodle.com/demos/simulate-nmr-and-ms/">https://web.chemdoodle.com/demos/simulate-nmr-and-ms/</a>
ACD/Labs iLabs - <a href="https://ilab.acdlabs.com/iLab2/">https://ilab.acdlabs.com/iLab2/</a> (limited online trial)
- Log in to post comments
Module 7: Assignment
1. Go to the NIST WebBook and download the following spectra in JCAMP-DX format
Mass spectra for each of the three isomers of nitrobenzoic acid
Gas phase IR spectra for each of the three isomers of nitroaniline
2. Go to ChemSpider and download the HNMR spectra of the three isomers of xylene in JCAMP-DX (can you find them?)
3. Go to the first JSpecView page above or download the JCAMP Viewer software for your OS. Import each of the spectra into the viewer you choose and export them to X,Y format.
4.Finally, import each of the X,Y data files into Excel with one sheet for the MS data, one sheet for the IR data, and another sheet for the NMR data. Plot the three IR spectra on one graph (so they are overlaid) and do the same for the MS and NMR data (so you end up with three graphs). Save the file.
5. Write a short paragraph comparing/contrasting for each of the thr
- Log in to post comments