Jordi Cuadros, (jordi.cuadros@iqs.url.edu)
Learning Objectives
- Get a basic understanding of Web APIs are accessed.
Introduction to web APIs
One common way to offer data to be used programmatically is through a web API. This learning object will be devoted to discuss how you access and use a web API.
What’s a web API?
An API (Application Programming Interface) is the set of elements that a programming library or service makes available to other programmers to be used remotely to access their services and data.
This API is called a web API when these functionalities are delivered via the HTTP protocol through the Internet.
Many web APIs exist, listings of public APIs can be found in https://www.publicapis.com/, https://any-api.com/ or https://www.programmableweb.com/.
How is a web API used?
In order to be able to get information from an online database, we will need to:
1.Send a query to the database through the Internet (HTTP protocol)
2.Understand the format in which the response will be received
3.Process this response to extract the desired information
Sending the query... - The HTTP protocol and the REST architecture
HTTP stands for HyperText Transfer Protocol and it comprises the set of rules that control the transfer of a resource between a web server and a web client (usually a browser).
The basic elements of an HTTP transaction are:
- A request message
- A response message (which is usually the resource being transferred)
The HTTP request message
The request message consists in some text data sent to an URL (Uniform Resource Locator). An URL is a way of specifying an address in the Internet.
The general structure for an URL consists of
scheme://[user:password@]domain:port/path?query_string#fragment_id
although in HTTP this commonly simplified to
http://domain/path or https://domain/path
The request message, which is usually handled by the HTTP client, has the following structure
- A first line which includes method, path and HTTP protocol version, usually
METHOD /path HTTP/x.x
- Several header lines
- A blank line
- And an optional message body
The HTTP request methods: GET and POST
Although other methods exist, the most common request methods are GET and POST.
In the GET method, any additional information required to specify the request is included in the URL (which means in the path included in the first line of the message). The parameters are included after the path and a question mark as pairs name=value. Parameters are separated by the ampersand symbol (&)
/path?name1=value1&name2=value2
In some cases, as we will see in PubChem API, the parameters are included in the path. This technique is called URL rewriting and shortens the URLs and makes it more usable.
/path/name1/value1/name2/value2
or even
/path/value1/value2
In POST methods, any additional information is not included in the URL but in the body of the message and is usually submitted via a web form. We won’t go further into this method since most of the chemistry APIs can be queried via GET calls.
To end this brief introduction to HTTP, note that some characters are either disallowed or have special meaning in URLs. When these characters appear they will need to be encoded; for example a space is encoded as %20, and a sharp (#) is %23. A reference can be found at http://www.w3schools.com/tags/ref_urlencode.asp.
The HTTP response message
As we have seen for the request, the response contains:
- A first line
- Several header lines
- A blank line
- And a message body which is the transferred resource
Two aspects of the response to be highlighted are the status code (included in the first line) and the content-type information (which is one of the headers).
HTTP status codes are indicated as a three-digit number
- 200 means transaction completed successfully
- 3xx means some type of redirection
- 4xx means a client-side error; e.g. 404 means resource not found
- 5xx means a server-side error; e.g. 500 means unexpected server error
The content type parameter indicates what data is included in the message body. Each type of resource is indicated with a standardized string (Internet media type; aka MIME type). For example, some common response formats are
- HTML file: text/html
- XML file: application/xml or text/xml
- CSV file: text/csv
- JSON file: application/json
- PNG image: image/png
Querying the databases
We have so far seen how the information is transmitted over the WWW. But how are databases accessed over the WWW? There are two main architectures used to access databases over the Net: SOAP and REST. Given its prevalence, we will center our discussion on the REST architecture.
Some major characteristics of the REST (REspresentational State Transfer) architecture are:
- It is usually implemented over HTTP.
- The queries are implemented as GET requests.
- The communication is stateless. Any query must contain all required information. Nothing is saved in the server.
- The query result is obtained as a response in a predefined format: XML, JSON, CSV, HTML, plain text...
Further reading
- http://code.tutsplus.com/tutorials/http-the-protocol-every-web-developer-must-know-part-1--net-31177
- http://code.tutsplus.com/tutorials/a-beginners-guide-to-http-and-rest--net-16340
- https://www.addedbytes.com/articles/for-beginners/url-rewriting-for-beginners/
- https://www.iana.org/assignments/media-types/media-types.xhtml
- http://www.restapitutorial.com/
Understanding the responses... - Common structured data filetypes
Common formats in which a web API may provide its response are HTML, XML, CSV, JSON and plain text. A short explanation of the first four types follows next. Feel free to skip it if you already know their characteristics.
HTML (HyperText Markup Language)
HTML is the standard language used to create web pages. When a web API outputs its result in HTML, this resource is usually intended to be viewed a browser. When no better option is available, HTML can be processed (parsed) to extract a specific information.
Likewise, any web page is published as HTML and so can be processed to extract any relevant information contained therein.
An example of HTML can be found here: http://kinetics.nist.gov/janaf/html/Cl-054.html
This the output of a search in the NIST-JANAF Thermodynamical Tables online database. The full HTML code can be read looking at the source of the page. A fragment is reproduced in Figure 1.
Figure 1. Part of the HTML code of a page
More information on HTML can be found at http://www.w3schools.com/html/.
XML (eXtensible Markup Language)
XML is a markup language designed to contain structured data in format easy to be read by computers and for humans. An example of an XML response can be found running the following query: https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/cid/1/property/MolecularFormula,MolecularWeight,InChIKey/XML. Figure 2 shows its response.
Figure 2. An XML response
More information on XML can be found at http://www.w3schools.com/xml/.
CSV (Comma Separated Values)
CSV, for Comma Separated Values, is a plain-text file format use to store tabular data. In practice, CSV is not a unique format but different variations depending on the implementation and the regional setup of the operating system.
CSV main features:
- Data is stored as plain text.
- Records are separated by new-line characters.
- Each record contains several fields. Columns (fields) are separated by a delimiting character (usually a comma, a semicolon or a tab).
- Each record must have the same number of fields.
CSV main variations:
- Any encoding may be used for the text file: ASCII, Latin-1, UTF-8...
- Values may be quoted or not.
- End-of-line character, delimiting character, quotes and escaping sequences may vary.
- There may be header lines.
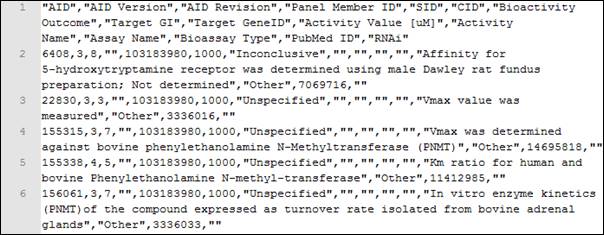
An example of a response in CSV is obtained when running the following query http://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/cid/1000,1001/assaysummary/CSV.
The response is shown in Figure 3.
Figure 3. Fragment of a CSV response
More information can be obtained at https://en.wikipedia.org/wiki/Comma-separated_values.
JSON (JavaScript Object Notation)
JSON, for JavaScript Object Notation, is a format that encodes data object as human-readable text, organized as pairs attribute-value (name:value). Its main features are being a quite compact notation with easy conversion to Javascript objects (eval(), JSON.parse()).
An example query is https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/name/aspirin/property/MolecularWeight/JSON. The response is shown in Figure 4.
Figure 4. A JSON response
More information can be found at http://www.json.org/.
Processing the responses... - Parsers and string functions
Ultimately the response will need to be processed to extract the desired information. This can be done using specific functions or parsers or through the use of some simple string manipulation functions.
Sometimes you want to parse XML or HTML using custom string functions for deleting whitespace, finding the position of certain characters within strings and for extracting substrings. These functions (plus many more) appear in all commonly used languages.
Table 1 summarizes some of these functions in the programming environments we will be discussing next week.
Table 1. Main string functions
|
Spreadsheet (Excel, Calc) |
Basic (VBA, Libre Basic) |
Javascript |
|
SEARCH, FIND |
InStr |
search, indexOf |
|
MID |
Mid |
substr, substring |
|
SUBSTITUTE |
Replace |
replace |
|
TRIM |
Trim |
trim |
|
CONCATENATE |
& (operator), |
concat |




Comments 23
Need Help!
Hi All,
I think this is mostly directed at Jordi, but thought I'd post it here, so the discussion is public. The jist of this email is that I am pulling data from one table, but would like to pull from more than one.
I have been working on cleaning up a sheet for webscraping WikiPedia Chemboxes, and the following link should be public.
https://docs.google.com/spreadsheets/d/11iWH9L8T7Kyue3UYg4ZUiYZ47LlBeNZPWGfIlEFxyrk/edit#gid=1454316265
If you click cell [C1] a drop down box created by data validation allows you to choose a property from the Wikipedia Chembox, which is then uploaded through the following query
=QUERY(IMPORTHTML(B2,"table",1),"select Col2 where Col1='"&$D$1&"'",0)
Note, as I had to use a filter in [D1] to pull the value out of the validation box in [C1], and essentially said that if the dropdown box says "density", set D1 to "Density", and then Density was used in the query.
the IMPORTHTML command takes data from the first table, and for the compounds in red, the chembox is the second table (the first is stuff from the wikipedia editors.)
How can I get the above script to pull from the first or second table?
In can use this and it works by itself, but not in the query.
={IMPORTHTML(URL, "TABLE", 7) ; IMPORTHTML(URL,"TABLE",8) ; IMPORTHTML(URL,"TABLE",9) }
I can bring in two tables, but I can't get that to work in the query.
I did go in and change the index for glucose in [C20] from 1 to 2, and now it works, but that is only a short term fix, and what is really needed is the ability to pick from the first or second table, as we have no idea when Wikipedia editors will be placing their message/comment tables on pages.
Any help is appreciated, and I was thinking this might morph into a project. For example, I think I have a strategy that can pull from multiple infoboxes
Google Sheet issues
Bob,
This sounds really cool! Unfortunately the behavior of the Google Sheet is not the same as you've described. I don't see a dropdown box in cell C1. Maybe that's because I don't have full permission to edit the page. The URL you posted says edit in there, but the permission I see at the top of the page says I only have permission to view.
Jennifer
updated permissions
OK, I think you can now edit. What I suggest/request, is that anyone who wants to edit make a duplicate of the sheet to that (tabs at the bottom), and play around, and don't worry about messing anything up.
I had an old version that used 2 columns for every chemical, which was much less elegant, but could take data from multiple tables in a page.
Cheers,
Bob
Cheers,
Bob
My current approach...
is here: https://docs.google.com/spreadsheets/d/15WjPrkY16HC_XgAuQWJ78F0-l_bj7DP1-H9d6d7MG8o/copy
I would say it is far from good but it does solve some issues: it looks for the infobox and it is a bit more resilient to the different names used for the properties in different compound pages.
Hope it helps.
Best,
Jordi
A slightly better option...
could be:
https://docs.google.com/spreadsheets/d/1Qw5UVxDSBiGKjxACs-PaUVjkZlZCCH1IM7brcfo9xS8/copy
I don't think I will be able to do more than this without scripting.
Best,
Jordi
A couple of Questions
Hi All, I think I am addressing Jordi, although anyone is welcome to chime in.
I tweaked your spreadsheet smiles2spectra to name2spectra and it is here,
https://docs.google.com/spreadsheets/d/1EGfeOYLYWT-wM_cHMN8zxDT1WoltOnDSjWRfFg0B35g/copy
I have two questions,
First, in this code,
=MID(IMPORTXML("https://chem.nlm.nih.gov/chemidplus/name/" & B1,"//link[@rel='canonical']/@href"),40,60)
I do not understand what is happening after the "&B1&", and I want to make a video explaining what we are doing. Can you explain?
Second, in cell 24 I made a jab at getting the jdx. Any advice? I would like to download the spectral data as two columns, can we do that?
Cheers,
Bob
additional question
Maybe I should mention to everyone, if you type a name into cell B1, it brings up an IR/UVVis/MS spectra (dictated by cell A5), and a jdx of the data.
Is there anyway we can lock some cells on a spreadsheet, while leaving others open? The ideal would be to lock all cells but B1. Can that be done?
Cheers,
Bob
First: XPath
For those wondering the SMILES2spectra spreadsheet is here https://docs.google.com/spreadsheets/d/1iKEXWUk4JQQdueklu8-PFbg-b0GNHYC9hIvoywEOY4I/copy.
The expression that is the second argument of the IMPORTXML function is an XPath expression. XPath is a computer language to pull specific data from an XML expression. More information on XPath can be found at:
https://www.w3.org/TR/xpath/
https://www.w3schools.com/xml/xpath_intro.asp
https://www.tutorialspoint.com/xpath/index.htm
"//link[@rel='canonical']/@href" reads search for a "link" tag wherever in the page such as it has an attribute "rel" which equals "canonical" and pull the value of its "href" attribute.
For benzene, it will pull the value "https://chem.nlm.nih.gov/chemidplus/rn/71-43-2" from line 106 of view-source:https://chem.nlm.nih.gov/chemidplus/name/benzene.
Does it make sense?
Cheers,
Jordi
Third: Protecting range and sheets in Google Sheets
This canbe done using the Data/Protected sheets and range...
I would suggest protecting the sheet Sheet1 but the ranges B1:D1 and B2, by using a warning on edit.
Cheers,
Jordi
Second: Import JCAMP-DX from NIST
Take a look to an improved version of the SMILES2Spectra sheet here:
https://docs.google.com/spreadsheets/d/1DG9fTdSkDE_f2DqEIW0ib2HOlW6p-BQH0TMqZbJUYBE/copy
JCAMP-DX import is in Sheet3. The chart works only when the data is in XYDATA format (usually IR spectra). More work needs to be done to process peaks or points.
MS Excel function equivalent to image() in googlesheet?
Hi, Jordi,
Does MS Excel have a function equivalent to image() used in googlesheet?
Sunghwan,
Not yet, but...
it can be emulated from a VBA function.
Take a look here: https://drive.google.com/file/d/0B5ln9gFZnHD4a0RtWWJCdWpKclE/view
Does it help?
Cheers,
Jordi
question on XPath
How would one load this page to a google sheet? https://pubchem.ncbi.nlm.nih.gov/rest/pug_view/annotations/heading/XML/?heading=Boiling+Point&response_type=display
I am assuming you place this in a cell (D52) https://pubchem.ncbi.nlm.nih.gov/rest/pug_view/annotations/heading/XML/?heading=Boiling+Point
And then something like =IMPORTXML(D52"//XPath_query")
But I do not know the XPath_query, or how to understand the Xpath_query
The information at this URL
so is there a way to upload
I can't see any easy way to
Here you have it
https://docs.google.com/spreadsheets/d/1PdBXwZFa_epfjMq4ZrFLiDxtIzJYtWtZtXew_Uc2XBY/copy
Question on Excel
another way
Simplified Google Sheets version
A simplified version following the approach Bob just suggested is available at https://docs.google.com/spreadsheets/d/1CJwl2Uz5ZQDsy0V7I0khi5IJM2mhMdMbLycaIrnIJyU/copy I also changed the URL to search only for the connectivity layers of the InChI. This way more chemicals are found although stereoisomerical and isotopical information is not preserved. Cheers, Jordi
Couple of Questions
For the xpath expression
"[@data-serp-pos='0']" means that the XML element has an attribute named data-serp-pos which value is '0'. I don't know what the attribute means but it seems to be related to the position of the link in the search results. Thus a value of '0' would mean the first result. Cheers, Jordi
For the text file format
What you have between the InChIKey and the next CID is a line-feed character. This character is not rendered in some text editors (e.g. Windows Notepad) but it is in others (e.g WordPad). In Unix/Linux, it means the start of a new line and this is the way it is interpreted in Excel. In Windows, a newline is indicated with two characters, carriage-return + line-feed and this is way it doesn't show in notepad. More detailed information can be found at https://en.wikipedia.org/wiki/Newline. Cheers, Jordi