2.1.1. The what and why of cheminformatics
The term cheminformatics contains its own definition: it is the science of how we can make use of chemical information. Most often, data and information about chemical compounds is either directly about molecular structure (for example, a 2D structural formula, or 3D atomic coordinates for a particular conformation of a compound) or is tied to a molecular structure (for example, physical properties of a compound, which you identify by its structural formula).
Cheminformatics involves storing, finding, and analyzing these structures using the data-processing power of computers. In order for (human) chemists to rely on insights from cheminformatics, however, they have to be able to understand the methods that computer programs employ, the way in which computers store and analyze chemical structure, and the results that they produce.
Therefore, cheminformatics depends upon the use of representations of molecular structures and related data that are understandable to both human scientists and to machine learning algorithms.
The origins of the field trace back to library science. Librarians have been organising books full of words for centuries, but the notion of indexing, sorting, searching and retrieving information using molecular structures is more novel. Professionals whom we would now refer to as cheminformaticians have spent decades developing ways to handle molecular data in context: making sure scientists can find out what they need to know, and match chemical compounds with literature publications, measured properties, synthetic procedures, spectra, and computational studies.
More recently, the field of cheminformatics has been adopted by the pharmaceutical industry. Since the 1980s, it has been standard procedure for large drug companies to manage their collections of potential drug compounds using computer software. These days, cheminformatics is usually associated with drug discovery, since it is the pharmaceutical industry that provides the demand for software that can keep track of literally millions of unique molecules that are available for R&D purposes.
2.1.2 Communicating with chemists vs. communicating with computers
How does communication between chemists differ from communication between a chemist and a machine? If one chemist was to recommend to another that a reaction should be performed using "chloroform" as a solvent for a reaction, this would generally be a successful exercise in communication. For all practical purposes, this word is understood by every chemist, and has no ambiguity.
If this fact were to be communicated to a machine, things start to get a little murky. Humans are quite accustomed to learning common facts, and after sufficiently many years spent studying at university, they tend to become very good at looking up information and inferring missing data. Software algorithms, however, are supremely literal. Because "chloroform" is a so-called trivial name, there is no formula for converting it into the actual chemical structure that it represents, and so a machine will not be able to participate in this exchange of information unless it has been explicitly instructed as to what the word means, using a format that it can work with.
A more descriptive way to communicate the composition that is chloroform is by chemical formula, in this case CHCl3. If this were handed over to a computer program, it would be a simple matter for it to understand that the substance being described is a molecule with 5 atoms: 1 carbon, 1 hydrogen and 3 chlorine. Assembling this into a molecule with bonds is a very simple matter, because 4 of the atoms are normally monovalent, and one of them is normally tetravalent. It is quite simple to create a software algorithm that can join the atoms together in the most obvious way, which also happens to be correct.
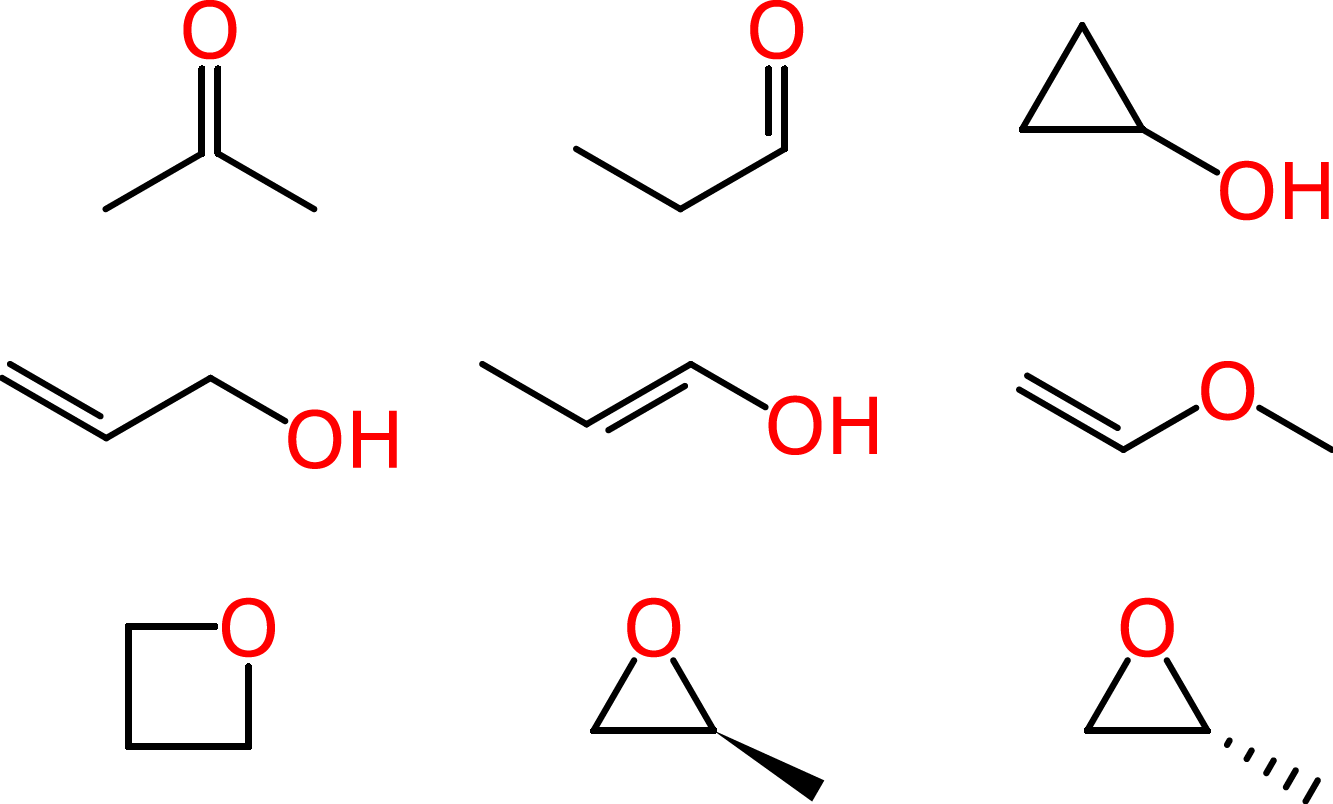
Beyond such tiny simple molecules, difficulties soon arise. Some of these ambiguities affect human chemists in the same way that they affect machines. Consider the molecular formula of C3H6O, which is associated with multiple reasonable structures, including a ketone, an aldehyde, a cyclic alcohol, oxygenated alkenes and cyclic ethers, one of which exists as two enantiomers:
Since systematic IUPAC names are made according to formalized rules, they could, in principle, be used by both humans and computers. However, as we have seen in the previous unit, IUPAC names are often quite difficult for chemists to read, let alone to write. Even when chemists more or less follow IUPAC rules in naming their substances, they often take shortcuts, creating their own abbreviated notations and shortened words. These may be well-understood by two scientists working in the same laboratory or in the same field, but they might lead to misunderstanding when it comes time to communicate findings with the larger community.
Interacting with a machine is a form of communication. It has a lot in common with two scientists speaking to each other about their research, but it has its differences, too. Because cheminformatics is first and foremost about gathering scientific facts that originate from scientists, it is essential to find a way for the humans who do the chemistry to communicate using a vernacular that the machine can understand.
In this case it may be helpful to think of machines and cheminformatics software as an extremely pedantic bureaucrat with a set of rules that are utterly rigid with no flexibility or room for interpretation, working exclusively according to a formulaic script. In this metaphor, if you provide everything that the bureaucrat requires, in the right order, then you will be served perfectly, and be on your merry way. If not, then your task will fail: either the bureaucrat will prevent you from moving to the next step because the script forbids it, or your inputs will be erroneously accepted and moved onto the next level, and fail later on.
As with our bureaucrat, in cheminformatics, you’re dealing with a system governed by strict rules, and these rules are not too difficult to define. If you know the rules, then you can make the system work for you. If you don't know them, your experience will result in frustration and failure, and there is no guarantee that you will even find out what went wrong until much later.
2.1.3. Structural formulas as chemical graphs
As we discussed in Part 1 of this module, usually, the most effective way to communicate with another chemist about the structure of a compound is to draw its structural formula.
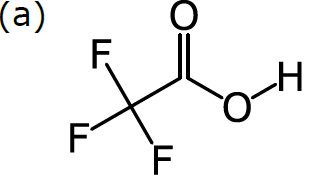
In order to do cheminformatics, we need to express chemical structure in a way that can be understood by machines as well as humans. It just so happens that structural formulas can be fairly directly mapped to a computer-friendly data structure. Structural formulas can be interpreted as a kind of graph: a set of nodes (in our case, atoms), certain pairs of which are linked by edges (in our case, bonds). For example, consider this structural formula for trifluoroacetic acid, CF3CO2H:
The diagram in (a) has no ambiguity, since all atoms are represented in a graph: there are 8 nodes, each of which is labelled according to the element, except for carbon (which is the default). Each of the 7 bonds is represented by an edge within the graph, and the bond order is represented by showing the number of lines. The way that this formula is drawn on paper is completely compatible with a data structure based on a labelled graph.
Such molecular graphs are typically stored in dedicated file formats designed for chemical information. (There are many to choose from; we’ll discuss the most popular ones a little later in this module and throughout the rest of this course.) This is good news, because cheminformaticians and computer scientists have come up with all kinds of clever data structures and algorithms for storing and analyzing datasets that can be represented as graphs.
However, just because structural formulas look like a graphs doesn’t mean that they always look like a chemically-meaningful graph – or like a graph with the same chemical meaning that the chemist who drew the structure intended. There is a long list of issues that need to be handled carefully in order to reliably encode all of the chemical information contained in a structural formula in a machine-readable manner. (Long enough to ensure that cheminformatics will continue to be a lively field of research for a very long time!)
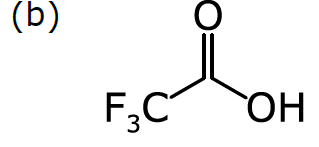
Consider a more common way of drawing trifluoroacetic acid:
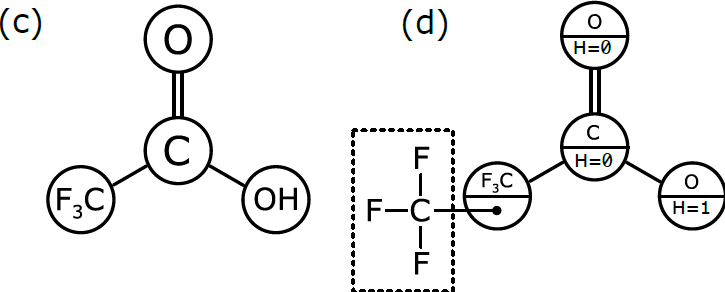
As we discussed in Part 1, chemists have become accustomed to condensing portions of structural formulas to make them easier to draw, read, and compare at a glance. The diagram (b) differs from the previous representation in that the carbon trisubstituted with fluoride has been collapsed into F3C, a single node (to use the terminology of graphs). So has the hydroxyl part of the acid (OH). A schematic form of the corresponding graph is shown in (c). In this case, the underlying graph has 4 nodes, not 8. The labels of these nodes are [O, C, F3C, OH]. Note that only two of these are elements from the periodic table. As shown, the structure represented by (c) is not a molecular graph, because some of its notes are labeled with something other than a symbol corresponding to an atom of a particular element.
In order to be interpreted by a computer as if it were a molecule, the molecular graph needs to be labelled in a slightly different way, which is represented schematically in (d). Here, we have annotated the graph nodes in a more systematic way.
Note that each node now has two labels. Three of the nodes are labeled so that the primary property is an atomic element, and the secondary property is the number of hydrogen atoms attached to it. These three node definitions are [O:H=0, C:H=0, O:H=1].
The fourth node, which represents a more complex group of atoms, is labeled slightly differently. Its primary property is the label F3C, which is displayed for the benefit of human chemists reading the structural formula. However, its secondary property is labeled with another graph that stores the configuration of atoms and bonds that makes up F3C. This is the underlying chemical information that the human chemist picks up easily when she sees “F3C” in a structural formula, but that needs to be described explicitly in order for the computer to properly understand and use it.
Graph (d) is the best of both worlds. The structure can easily be displayed in the way that chemists expect to see it, but it can also be easily interpreted by a computer algorithm, because the definition follows rules that allow the full atomic structure to be reassembled behind the scenes.
Most importantly, always remember that just because a chemical structure has been digitized and stored on a computer does not mean that the information can be used by cheminformatics. In fact, most of the chemical structures that have been generated by scientists and put on computers are available not in a graph-based file format, but in a nonchemical graphics format. (There are two main types of such nonchemical image files: bitmapped and vector graphics. Neither is of much use for cheminformatics purposes.)
In order to make use of any of the capabilities that cheminformatics offers, the molecules involved must be represented as a molecular graph, rather than such generic print-ready forms. Furthermore, the molecular graph must be sufficiently well defined that an algorithm can use the information to piece together the complete structure. Every single atom and bond must be present and accounted for, in some way or another.
Different data structures do so in different ways, each of which has its advantages and disadvantages. For example, some approaches try to keep the representation as similar as possible to the human-friendly version (e.g. Kekulé forms for aromatic rings). Others favor a selection of properties that is more similar to the quantum wave-function of the molecule (e.g. use of resonance/aromatic bonds to denote equivalence).
2.1.4. Three dimensions of machine-readable chemical representation
A chemical structure representation contains two kinds of information: explicit and implicit. [H1] Explicit information is what’s directly represented in a data structure. Implicit information is what you (or a computer) can figure out from a data structure, given some knowledge of general principles and a little bit of work.
In general, data structures that contain less explicit information are more simple and compact, but they require more computation to draw chemical conclusions from them. Data structures that contain more explicit information take up more space and are at greater risk of containing inconsistencies, but they can be more quickly analyzed in a wider variety of ways.
In its essence, a molecular graph is a description of the relationships among a set of atoms that makes up a molecule. Like the condensed formulas and systematic names that we talked about in the first part of this module, a molecular graph can therefore be expressed in a one-dimensional (1D), or linear form. Such linear data structures are particularly well-suited to many of the best known cheminformatics applications, such as determining:
- whether molecules are the same.
- how similar they are, according to some metric.
- whether one molecular entity is a substructure of another.
- whether two molecules are related by a specific transformation.
- what happens when molecules are cut into pieces and grafted together at different positions.
In these and other applications of cheminformatics, linear representations have key advantages, especially when you’d like to handle huge numbers of structures very quickly (e.g. searching a large database).
However, chemists most frequently think about structure in 2D, and molecules actually exist in 3D physical space. Therefore, some data structures add 2D or 3D “diagram-like” coordinates for each atom.
In the 2D case, these 2D coordinates can be used to infer chemical information, such as the E/Z geometry of alkene-like double bond, or the cis/trans isomerism of ligands in a square planar metal complex, or substituents on a cyclic alkane. However, these coordinates are also descriptions for how exactly the structural formula is drawn. If you’re interested in communicating with other chemists as well as with a computer, you may wish to draw a structural formula in some particular way, to emphasize a certain bond or group. By storing the 2D coordinates of each atom, some data structures are designed to keep track of these choices, in order to facilitate human communication as well as machine analysis.
Other data structures are designed to represent the real 3D shape of a molecule. The molecular structure complete with a 3D (x,y,z) coordinate for each atom is often referred to as a conformation or a model. Such a data structure makes a scientific claim that the particular coordinates that it contains represent the molecule's actual shape, whether it be in solution, in a vacuum, or in the binding site of a protein. This opens up a whole new domain of computational chemistry, especially since most molecules have some flexibility. Even if a given conformation is the most stable, there often a number of competing shapes that must also be considered.
These coordinates may be determined experimentally (typically via x-ray crystallography). They may also be calculated, using force-fields (which treat atoms and bonds like "balls and springs" using classical physics), quantum chemistry (which solve the Schrödinger equation by various approximations), molecular dynamics (which model motion over time) or composite models such as docking (which are designed for specific environments, like peptide binding sites).
Keep in mind that even a data structure that provides 3D coordinates **may not tell you** where those coordinates come from. Knowing how a particular set of coordinates was determined is crucial to making intelligent use of it for cheminformatics purposes.

Comments 2
I am confused on how a
Interpreting chemical graphs