2.4. Exercises (Module 4b)
2.4.1. Exercise 1 NOTE* You may want to download the MS Word Document at the top of the module homepage
Fill in the following table using both ChemSpider and PubChem and indicate where answers may differ. Use the new compound identity search in PubChem (click on the hexagon for structural formula, https://pubchem.ncbi.nlm.nih.gov/search/#collection=compounds&query_type=structure&query_subtype=identity)
|
molecular formula |
Structural formula |
Systematic name |
SMILES |
InChI |
CAS RN |
|
|
|
|
CC(=C)C=C |
|
|
|
|
|
|
|
|
|
|
|
|
Ammonium acetate |
|
|
|
|
C6H6O |
|
|
|
|
|
|
|
|
|
|
|
105-53-3 |
2.4.2. Exercise 2
Which of above form(s) of notation is/are preferable for:
- Ordering a specific compound from a supplier
- Identifying an important compound in a journal article
- Sorting a list of a bunch of compounds into chemically-meaningful groups
2.4.3. Exercise 3
Search PubChem for lactic acid (racemic) and its two enantiomers, shown in Module 4a, Figure III.a-e and in section 2.2.2 above.
- Paste the urls, IUPAC names, and CAS numbers that you find below.
- You should see something strange in your answer to Part b). What is it?
- Luckily, the makers of PubChem have followed some of the best practices that we’ve outlined above. What have they done that can help you get to the bottom of the inconsistency that you’ve discovered in parts (b-c)?
- Follow the clue that PubChem left you as to the source of the discrepancy and describe what might have happened with the data coming into Pubchem. (If you aren’t sure, what to do, click http://www.drugbank.ca/drugs/DB03066 and http://www.drugbank.ca/drugs/DB04398).

Comments 20
Alternative Word Processors
You may find interesting to
Registry number
Getting CAS
CAS numbers
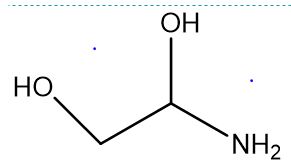
1-aminoethane-1,2-diol questions
Fun Fact
removal of a compound
removal of a compound
Removal of a Compound
Deprecation of records from ChemSpider
Question on Provenance
Re: Question on Provenance
Chemistry Electronic Lab Notebooks
1-aminoethane-1,2-diol
Re: chemspider search
Pulling structures
Downloading multiple structures from SMILES or InChIs
Question 2.4.3
Re: Question 2.4.3